

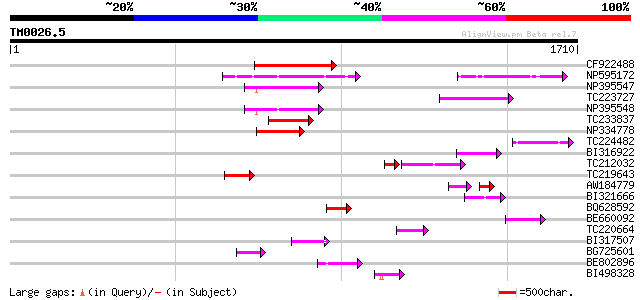
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.5
(1710 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922488 209 1e-53
NP595172 polyprotein [Glycine max] 207 3e-53
NP395547 reverse transcriptase [Glycine max] 160 4e-39
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 151 2e-36
NP395548 reverse transcriptase [Glycine max] 150 4e-36
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 145 1e-34
NP334778 reverse transcriptase [Glycine max] 137 4e-32
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 110 4e-24
BI316922 96 1e-19
TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 70 3e-18
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 91 4e-18
AW184779 59 2e-15
BI321666 70 1e-11
BQ628592 67 5e-11
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 64 7e-10
TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial... 62 2e-09
BI317507 62 3e-09
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 62 3e-09
BE802896 61 4e-09
BI498328 59 2e-08
>CF922488
Length = 741
Score = 209 bits (531), Expect = 1e-53
Identities = 106/249 (42%), Positives = 163/249 (64%)
Frame = +3
Query: 737 VIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAE 796
V+K++GK+ +C+D+RDLN A+PKD++ +P ++VD+T S +DG+SGYNQI IA
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 797 EDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKS 856
ED+ KT F GT+ + M FGLKN GATYQR M +F D + ++VY+DD++VKS
Sbjct: 183 EDMEKTTFIT--LWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKS 356
Query: 857 PSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILD 916
+ ++HL +LRK F R+RKY L++NP KC F V + L F+ +GIE++ NK K IL+
Sbjct: 357 RTEEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILE 536
Query: 917 TSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELK 976
+ P ++KQ+Q LG++N++ RFI+ L + + L K +W+ + AF+ +K
Sbjct: 537 MAKPHTEKQVQGFLGRLNYIVRFIS*LIATCEPL--FILLCKNQFVKWDHDC*VAFERIK 710
Query: 977 VYLSSPHVM 985
L +PHV+
Sbjct: 711 QCLINPHVL 737
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 207 bits (527), Expect = 3e-53
Identities = 138/415 (33%), Positives = 210/415 (50%)
Frame = +1
Query: 642 DEEKDATTEWQNEMVKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRF 701
D KD T E+ LL + FA P +D +P+K+ PVK P R+
Sbjct: 1645 DTLKDLPTNIDPELAILLHTYAQVFAVPASLPPQREQDHA---IPLKQGSGPVKVRPYRY 1815
Query: 702 HPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDE 761
+I++ I+ +L I+ + L ++ V KK+G R C D+R LNA T KD
Sbjct: 1816 PHTQKDQIEKMIQEMLVQGIIQPSNSPFSLP-ILLVKKKDGSWRFCTDYRALNAITVKDS 1992
Query: 762 YHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFG 821
+ MP + ++D G +Y S LD SGY+QI + ED KTAFR G YEW+VMPFG
Sbjct: 1993 FPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHH--GHYEWLVMPFG 2166
Query: 822 LKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMN 881
L NA AT+Q +MN IF + F+ V+ DDI++ S S DHL HL + ++++ L
Sbjct: 2167 LTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFAR 2346
Query: 882 PLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIA 941
KC+FG D+LG V G+ + K +A+LD P + KQL+ LG + RRFI
Sbjct: 2347 LSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIK 2526
Query: 942 NLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISAT 1001
+ + PL L ++D+F W E + AF +LK ++ V++ P +P L A+
Sbjct: 2527 SYANIA---GPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDAS 2697
Query: 1002 DGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYI 1056
+G++L Q I Y S+ L + + + L + + K ++Y+
Sbjct: 2698 GIGVGAVLGQNG-----HPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYL 2847
Score = 97.4 bits (241), Expect = 5e-20
Identities = 91/339 (26%), Positives = 144/339 (41%), Gaps = 7/339 (2%)
Frame = +1
Query: 1350 HDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSII 1409
H G H AGI + YWP + +D Y + C Q+ +PA L +
Sbjct: 3235 HSSPIGGH-AGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPL- 3408
Query: 1410 KPWPFRGW---ALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQ-NVTQDTVIDFIQ 1465
P P + W A+D I + +S I+V ID TK+ IPL+ + V +
Sbjct: 3409 -PIPQQVWEDVAMDFITGLP--NSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFM 3579
Query: 1466 NHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLIS 1525
+HIV G+P S+ +D+ VF + G L S+ Y+ Q++GQ E NK L
Sbjct: 3580 SHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEM 3759
Query: 1526 LIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCRIQR 1585
++ PK W + L + Y + + G TPFR YG+E P + Q+C I
Sbjct: 3760 YLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGRE---PPTLTRQACSIDD 3930
Query: 1586 QEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVWKV 1645
E+ ++L + D LTR + + + +KK SF GD V
Sbjct: 3931 PAEV---------REQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVK 4083
Query: 1646 VLPVDKRD---KRYGKWAPNWEGPFTVEKILLNNAYSIK 1681
+ P + ++ K + + GPF V + + AY ++
Sbjct: 4084 LQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLE 4200
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 160 bits (405), Expect = 4e-39
Identities = 95/255 (37%), Positives = 133/255 (51%), Gaps = 18/255 (7%)
Frame = +1
Query: 709 IKEEIERLLKCKFIRTARYVDWLANVVPVIKKNG------------------KMRVCIDF 750
+++E+ +LL+ I W++ V V KK G + R+CID+
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 751 RDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGAL 810
R LN AT KD Y +P + M+ A + LDGYSGYNQI + +D KTAF CP ++
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 811 GTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSF 870
Y MPFGL NA T+QR M IF D +E ++V++DD S + LA+L K
Sbjct: 361 FAYRR--MPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVL 534
Query: 871 ERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLL 930
+R K L +N KC F V G LG + K+GIE+ K K I PP + K + S L
Sbjct: 535 QRCEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFL 714
Query: 931 GKINFLRRFIANLSE 945
G + F RRFI + ++
Sbjct: 715 GHVGFYRRFIKDFTK 759
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 151 bits (382), Expect = 2e-36
Identities = 80/223 (35%), Positives = 116/223 (51%)
Frame = +1
Query: 1297 DYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGA 1356
+YL R + A + + G+ L+K+N D L+C+ +A I VH+G G
Sbjct: 175 EYLPEIADNDKRTLRRLAAGFFMSGSILYKRNHDMKPLRCVDAREANHMIEEVHEGSFGT 354
Query: 1357 HQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRG 1416
H G M + R G YW T+ DC + + C Q A + P L+ + PWPF
Sbjct: 355 HANGHAMARKILRAGYYWLTMESDCCVHVRKCHKCQAFADNVNAPPHPLNVMSSPWPFSM 534
Query: 1417 WALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPE 1476
W +D+IG I P +S H++I+VAIDYFTKWVEA +V + V+ FI+ I+ R+GLP
Sbjct: 535 WGIDVIGAIEPKASNGHRFILVAIDYFTKWVEAASYTDVMRGVVVRFIKKEIICRYGLPR 714
Query: 1477 SLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAA 1519
+ TD GT + + E + I+ N TPY + N VE A
Sbjct: 715 KIITDNGTNLNNKMMGEICEEFKIQHHNPTPYRPKMN*AVEVA 843
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 150 bits (380), Expect = 4e-36
Identities = 88/255 (34%), Positives = 133/255 (51%), Gaps = 18/255 (7%)
Frame = +1
Query: 709 IKEEIERLLKCKFIRTARYVDWLANVVPVIKKNG------------------KMRVCIDF 750
+++E+ +LL+ I W++ V+ V KK G ++CID+
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 751 RDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGAL 810
R LN AT KD + +P + M++ AGH Y LD Y GYNQI + +D K AF CP
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCP--F 354
Query: 811 GTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSF 870
G + + +PFGL NA T+Q M IF D +E ++V++DD V PS + L L
Sbjct: 355 GVFAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVL 534
Query: 871 ERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTSPPTSKKQLQSLL 930
+R + L +N KC F V G LG + +GIE+++ K I PP++ K ++S L
Sbjct: 535 QRCVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFL 714
Query: 931 GKINFLRRFIANLSE 945
G+ F RRFI + ++
Sbjct: 715 GQARFYRRFIKDFTK 759
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 145 bits (366), Expect = 1e-34
Identities = 71/134 (52%), Positives = 94/134 (69%)
Frame = +2
Query: 781 SLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDF 840
S +DG+SGYNQI +A EDV KT F GT+ + VM FGLKN GATYQR M +FHD
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVT--LWGTFSYRVMAFGLKNTGATYQRAMVALFHDM 178
Query: 841 IETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVH 900
+ ++VY+DD++ KS + +HL +L K F R++KY LK+NP KC FGV +G LGF+V
Sbjct: 179 MHKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVS 358
Query: 901 KKGIEINKNKAKAI 914
+KGIEI+ K KA+
Sbjct: 359 QKGIEIDPEKVKAL 400
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 137 bits (345), Expect = 4e-32
Identities = 63/145 (43%), Positives = 97/145 (66%)
Frame = +3
Query: 745 RVCIDFRDLNAATPKDEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 804
R+C+D+RDLN A+PKD + +P ++++ + A S +DG+SGYNQI +A ED+ KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 805 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLA 864
GT+ + VM FGLKN GATY R M +F D + ++ Y+D+++ KS ++HL
Sbjct: 183 IT--LWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLV 356
Query: 865 HLRKSFERMRKYGLKMNPLKCAFGV 889
+L+ F ++RKY L++NP KC FG+
Sbjct: 357 NLQNLFGQLRKYRLRLNPRKCVFGL 431
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 110 bits (276), Expect = 4e-24
Identities = 66/184 (35%), Positives = 98/184 (52%)
Frame = +1
Query: 1517 EAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEV 1576
EAANK + +I+K + K WH+ L L YR S + +TGATPF L YG EAVLP EV
Sbjct: 1 EAANKNIKKIIQK-MTVSYKDWHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPFEV 177
Query: 1577 YLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSF 1636
+ S RI + + ++ D+L ++ +RL+A+ + R+ A++KKV + F
Sbjct: 178 EVPSLRILAESGLKESEWAQTRYDQLNLIEGKRLTAMSHGRLYQQRMKSAFDKKVCLRKF 357
Query: 1637 MPGDYVWKVVLPVDKRDKRYGKWAPNWEGPFTVEKILLNNAYSIKELGGRNRQMTVNGKY 1696
GD V K + K + GKWAPN+EGPF V++ A + + G +N
Sbjct: 358 HEGDLVLKKMSHAVKDHR--GKWAPNYEGPFVVKRAFSGGALVLTNMDGEELPSPMNSDV 531
Query: 1697 LKTY 1700
+K Y
Sbjct: 532 VKRY 543
>BI316922
Length = 405
Score = 96.3 bits (238), Expect = 1e-19
Identities = 50/133 (37%), Positives = 74/133 (55%)
Frame = +3
Query: 1349 VHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSI 1408
+H G+CG H M + R G Y T+ K C EY K C++ + I H+ ELH+I
Sbjct: 6 MHRGICGMHSKSQLMTTRVLRVGYY**TMRKYCTEYVKKCEEC*KFGNISHLLVEELHNI 185
Query: 1409 IKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHI 1468
+ PWPF +D++ P S RQ KY++V ID FTKW+E + ++ V F+ +I
Sbjct: 186 VAPWPFAI*GVDILRPF-PLSKRQVKYLLVGIDQFTKWIETEHIAIISIANVRKFV*RNI 362
Query: 1469 VYRFGLPESLTTD 1481
V FG+P +L +D
Sbjct: 363 VC*FGIPNTLISD 401
>TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (3%)
Length = 803
Score = 69.7 bits (169), Expect(2) = 3e-18
Identities = 56/200 (28%), Positives = 89/200 (44%), Gaps = 6/200 (3%)
Frame = +2
Query: 1181 LEILIAFGAKNVVIKGDSELVIKQLTKEYKCVSENLARYYTKANNLLAKFDEARLSHVSR 1240
++ I K + + GDS LVI QL E + NL Y L FDE HV+
Sbjct: 206 VQAAIDSNVKLLKVYGDSALVIHQLRGECETRDPNLIPYQAYIKELAGFFDEISFHHVA* 385
Query: 1241 VDNQEANELAQIASGYMVDKCRLKELIEVKEKLNLSDLNILVIDNMAPNDWRKPIVDYLQ 1300
+NQ A+ LA + S + + IE + + + LV + W I Y++
Sbjct: 386 EENQMADALATLVSMFQLTPHGDLPYIEFRCRGRPAHC-CLVEEERDGKPWYFDIKRYVE 562
Query: 1301 N---PVGTTD---RKTKYRAMSYVIMGNELFKKNVDGTLLKCLSEDDAFIAISAVHDGLC 1354
+ P+ +D R+ + A + + G+ L+K+N D LL C++ + + VH+G
Sbjct: 563 SKEYPLEASDNDKRRKRRLAAGFFMSGSILYKRNHDMVLLHCVNGKEVENMLGEVHEGSF 742
Query: 1355 GAHQAGIKMKWILFRQGMYW 1374
G H G M + R G YW
Sbjct: 743 GTHSNGHAMARKILRAGYYW 802
Score = 42.4 bits (98), Expect(2) = 3e-18
Identities = 20/47 (42%), Positives = 29/47 (61%)
Frame = +1
Query: 1130 WKLFFDGSSHKNGTGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEA 1176
W ++FD +S+ G G+G +VSP F R+ +C+NN AEYEA
Sbjct: 52 WIVWFDRASNVLGHGVGAILVSPDNQCIPFTTRLGFDCTNNMAEYEA 192
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 90.9 bits (224), Expect = 4e-18
Identities = 39/90 (43%), Positives = 61/90 (67%)
Frame = +3
Query: 648 TTEWQNEMVKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLV 707
T + E++ LL++++D FAW Y +MPGLS D+V+ +LP+ + PVKQ RR P+ +
Sbjct: 1047 TAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNPECSPVKQKLRRMKPETSL 1226
Query: 708 KIKEEIERLLKCKFIRTARYVDWLANVVPV 737
KIKEE+++ F+ ARY W+AN+VP+
Sbjct: 1227 KIKEEVKK*FDAGFLAVARYPKWVANIVPI 1316
Score = 33.1 bits (74), Expect = 1.1
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +1
Query: 545 WIHGVGAVPSTLHQRI 560
WIH VG VPSTLHQ++
Sbjct: 4 WIHSVGVVPSTLHQKL 51
>AW184779
Length = 432
Score = 58.5 bits (140), Expect(2) = 2e-15
Identities = 25/45 (55%), Positives = 33/45 (72%)
Frame = +3
Query: 1417 WALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVI 1461
W +D+IG I P +S H +I+VAIDYFTKWVEA+ +VT+ VI
Sbjct: 294 WGIDVIGAIEPKASNGHHFILVAIDYFTKWVEAVSYASVTRSVVI 428
Score = 43.5 bits (101), Expect(2) = 2e-15
Identities = 24/71 (33%), Positives = 34/71 (47%)
Frame = +1
Query: 1322 NELFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDC 1381
N L+K+N D LL+C+ +A + VH+G G H M + R G YW T+ DC
Sbjct: 10 NILYKRNHDMVLLRCVDAREAEQMLVEVHEGSFGTHANIHAMAQKILRVGYYWLTMESDC 189
Query: 1382 MEYDKGCQDFQ 1392
+ C Q
Sbjct: 190 CIHVWKCHKCQ 222
>BI321666
Length = 430
Score = 69.7 bits (169), Expect = 1e-11
Identities = 39/122 (31%), Positives = 59/122 (47%)
Frame = +2
Query: 1373 YWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQ 1432
Y P+I KD + + C QR + LH+I++ F W +D +G P S +
Sbjct: 32 YLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFVGPFPPSFSNE 211
Query: 1433 HKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVA 1492
YI+V +DY +KWVEA+ Q VI F++ I R G+P L + G+ +A
Sbjct: 212 --YILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLIDNGGSHLCNA*LA 385
Query: 1493 SF 1494
F
Sbjct: 386 RF 391
>BQ628592
Length = 423
Score = 67.4 bits (163), Expect = 5e-11
Identities = 30/76 (39%), Positives = 50/76 (65%), Gaps = 1/76 (1%)
Frame = -1
Query: 956 LKKEDAFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDED 1015
L K A W + +Q+AF+++K L++P V+ PP+ G+P LY++ D ++G +L Q D+
Sbjct: 423 LPKNQAILWNSNYQEAFEKIKQSLANPSVLMPPVTGRPFLLYMTMLDESMGCVLVQHDDS 244
Query: 1016 -SKERAIFYLSRVLND 1030
KE+AI+YLS+ D
Sbjct: 243 GKKEQAIYYLSKKFTD 196
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 63.5 bits (153), Expect = 7e-10
Identities = 35/119 (29%), Positives = 59/119 (49%)
Frame = -3
Query: 1496 ESWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKE 1555
+ +G+ STPY+ Q NGQ E +N+ + +++K V K W L LWA+R + K
Sbjct: 361 KKYGVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTAYKA 182
Query: 1556 ATGATPFRLAYGQEAVLPAEVYLQSCRIQRQEEIPSEDYWNMMLDELVNLDEERLSALD 1614
G +P+R+ +G+ LP E+ ++ + + +L LDE RL A +
Sbjct: 181 PIGMSPYRVVFGKACHLPVEIEHKAYWAVKTCNFSMDQAGEERKLQLSELDEIRLEAYE 5
>TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (31%)
Length = 724
Score = 62.4 bits (150), Expect = 2e-09
Identities = 35/98 (35%), Positives = 55/98 (55%), Gaps = 4/98 (4%)
Frame = +1
Query: 1168 SNNEAEYEALISGLEILIAFGAKNVVIKGDSELVIKQLTKEYKCVSENLARYYTKANNLL 1227
+NN AEY A+I G++ + G + I+GDS+LV Q+ +K +ENL+ Y A L
Sbjct: 64 TNNAAEYRAMILGMKYALKKGFTGIRIQGDSKLVCMQIDGSWKVKNENLSTLYNVAKELK 243
Query: 1228 AKFDEARLSHVSRVDNQEANELA----QIASGYMVDKC 1261
KF ++SHV R N +A+ A +A G + ++C
Sbjct: 244 DKFSSFQISHVLRNFNSDADAQANLAINLADGQVQEEC 357
>BI317507
Length = 359
Score = 61.6 bits (148), Expect = 3e-09
Identities = 33/114 (28%), Positives = 58/114 (49%)
Frame = -1
Query: 851 DIVVKSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNK 910
+I++ SP H+ HL + ++K L N KC F ++LG V+ K + ++ NK
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 911 AKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRW 964
K++++ P + K++ S L + R+FI + + PL L K D F+W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAP--RPLTDLTKNDGFKW 18
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 61.6 bits (148), Expect = 3e-09
Identities = 36/89 (40%), Positives = 47/89 (52%)
Frame = -3
Query: 684 KLPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGK 743
KL I D K V Q R+ + + +E+ +L FIR Y L +VV V K NGK
Sbjct: 274 KLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNGK 95
Query: 744 MRVCIDFRDLNAATPKDEYHMPVAEMMVD 772
R+C D+ DLN A PKD Y +P + M D
Sbjct: 94 WRICTDYIDLN*ACPKDAYPLPNIDHMTD 8
>BE802896
Length = 416
Score = 61.2 bits (147), Expect = 4e-09
Identities = 45/137 (32%), Positives = 69/137 (49%)
Frame = -2
Query: 928 SLLGKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEAEHQKAFDELKVYLSSPHVMAP 987
S LG F RRFI + + S LL+ KE F + + + AFD LK L + ++
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQ--KEVEFDFNDKCK*AFDCLKRALITTPIIQA 242
Query: 988 PIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYF 1047
P P +L A++ +G +LAQ+ D R I+Y SR L+ A+ YT EK L + F
Sbjct: 241 PDWTAPFELMCDASNYALGVVLAQK-IDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVF 65
Query: 1048 SCVKLKYYIKPIDVMVF 1064
+ K Y+ ++V+
Sbjct: 64 ALEKFHSYLLGTRIIVY 14
>BI498328
Length = 335
Score = 58.9 bits (141), Expect = 2e-08
Identities = 39/108 (36%), Positives = 53/108 (48%), Gaps = 18/108 (16%)
Frame = +1
Query: 1101 KAVKGQAIADFLVDHTL----------PKEIV--------TYVGIQPWKLFFDGSSHKNG 1142
KAVKG A+AD+L L P E + T+ I W + FDG+S+ G
Sbjct: 10 KAVKGSALADYLAQ*PLQGYRPMHPEFPDEDIMALFEEKRTHEDINKWIVCFDGASNALG 189
Query: 1143 TGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGLEILIAFGAK 1190
G+G +VSP F R+ +C+NN AEYEA G++ I F K
Sbjct: 190 HGVGAVLVSPDDQCIPFTARLGFDCTNNMAEYEACALGVQAAIDFDVK 333
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 74,816,796
Number of Sequences: 63676
Number of extensions: 1061450
Number of successful extensions: 5824
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 5550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5787
length of query: 1710
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1600
effective length of database: 5,635,272
effective search space: 9016435200
effective search space used: 9016435200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0026.5


