

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
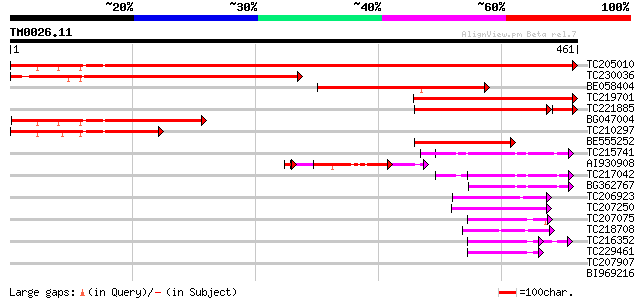
Query= TM0026.11
(461 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205010 UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate reductas... 738 0.0
TC230036 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosul... 357 8e-99
BE058404 similar to GP|18252504|gb adenosine 5'-phosphosulfate r... 256 1e-68
TC219701 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosul... 218 3e-57
TC221885 similar to UP|Q6V3B0 (Q6V3B0) Adenosine 5' phosphosulfa... 182 4e-52
BG047004 similar to GP|18252504|gb adenosine 5'-phosphosulfate r... 152 3e-37
TC210297 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosul... 121 6e-28
BE555252 113 2e-25
TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like prot... 57 1e-08
AI930908 similar to GP|18252504|gb| adenosine 5'-phosphosulfate ... 56 1e-08
TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like pro... 54 1e-07
BG362767 51 1e-06
TC206923 similar to UP|Q8LAM5 (Q8LAM5) Protein disulfide isomera... 47 2e-05
TC207250 similar to UP|Q9LW75 (Q9LW75) Dbj|BAA81871.1, partial (... 47 2e-05
TC207075 similar to UP|Q84XU4 (Q84XU4) Protein disulphide isomer... 46 3e-05
TC218708 similar to GB|AAF40463.1|7211992|F13M7 Strong simialrit... 45 7e-05
TC216352 protein disulfide isomerase-like protein [Glycine max] 44 1e-04
TC229461 similar to UP|Q84XU4 (Q84XU4) Protein disulphide isomer... 42 6e-04
TC207907 similar to UP|Q96313 (Q96313) AT.I.24-13 protein, parti... 39 0.005
BI969216 weakly similar to GP|28140231|gb protein disulphide iso... 37 0.014
>TC205010 UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate reductase, complete
Length = 1921
Score = 738 bits (1905), Expect = 0.0
Identities = 369/472 (78%), Positives = 413/472 (87%), Gaps = 11/472 (2%)
Frame = +2
Query: 1 MALAVTCSISISSSSSSSSTF-----QSSQPKFSQIASIRVSE--IPHGGGVNASQRRSF 53
MALAV+ + S S+++++SS+F SS K QI S R E + VN +QRRS
Sbjct: 44 MALAVSTTSSSSAAAAASSSFFSRLGSSSDAKAPQIGSFRFPERSLVSSVVVNVTQRRSL 223
Query: 54 VKP----PQRSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGN 109
V+P PQR+ DSI PLAATIVA +V E E+++++Q+A DLENASPL+IMD ALEKFGN
Sbjct: 224 VRPLNAEPQRN-DSIVPLAATIVAPEV-EKEEEDFEQIAKDLENASPLEIMDRALEKFGN 397
Query: 110 HIAIAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPD 169
IAIAFSGAEDVALIEYA LTGRP+RVFSLDTGRLNPETY+FFD VEKHYGIHIEYMFPD
Sbjct: 398 DIAIAFSGAEDVALIEYAHLTGRPYRVFSLDTGRLNPETYKFFDAVEKHYGIHIEYMFPD 577
Query: 170 AVEVQGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVP 229
AVEVQ LVR+KGLFSFYEDGHQECCRVRKVRPLRRALKGL+AWITGQRKDQSPGTRSE+P
Sbjct: 578 AVEVQALVRTKGLFSFYEDGHQECCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIP 757
Query: 230 VVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCT 289
VVQVDPVFEG+DGG+GSLVKWNPVANV G DIWSFLRTM+VPVNSLHS+GY+SIGCEPCT
Sbjct: 758 VVQVDPVFEGLDGGIGSLVKWNPVANVNGLDIWSFLRTMDVPVNSLHSQGYVSIGCEPCT 937
Query: 290 RAVLPGQHEREGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTATVADIFNTQ 349
R VLPGQHEREGRWWWEDAKAKECGLHKGN+K + A+LNGNG + NG+ATVADIFN+Q
Sbjct: 938 RPVLPGQHEREGRWWWEDAKAKECGLHKGNIKHEDAAQLNGNGASQANGSATVADIFNSQ 1117
Query: 350 NVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFR 409
+VVSLSR+GIENL KLENRKEPWLVVLYAPWC +CQAMEESYVDLA+KLA SGVKV KFR
Sbjct: 1118DVVSLSRSGIENLAKLENRKEPWLVVLYAPWCRFCQAMEESYVDLAEKLAGSGVKVAKFR 1297
Query: 410 ADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNALR 461
ADG+QKE+AK EL LGSFPTI+ FPKHSS+PIKYPSE RDVDSL AFVNALR
Sbjct: 1298ADGDQKEYAKTELQLGSFPTILLFPKHSSQPIKYPSEKRDVDSLTAFVNALR 1453
>TC230036 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate
reductase, partial (41%)
Length = 750
Score = 357 bits (915), Expect = 8e-99
Identities = 186/244 (76%), Positives = 207/244 (84%), Gaps = 6/244 (2%)
Frame = +3
Query: 1 MALAVTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVN--ASQRRSFVKP-- 56
MALA T SIS + +STF SS+PK QI SIR+SE P GG VN SQRRS VKP
Sbjct: 36 MALAFTSSIS-----APTSTFPSSEPKLPQIGSIRISERPIGGAVNFNLSQRRSLVKPVN 200
Query: 57 --PQRSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAIA 114
P R +DSI PLAAT + + +ET++++++Q+A DL+NASPL+IMD AL+KFGN IAIA
Sbjct: 201 AEPPR-KDSIVPLAATTIVASASETKEEDFEQIASDLDNASPLEIMDRALDKFGNDIAIA 377
Query: 115 FSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEVQ 174
FSGAEDVALIEYA+LTGRPFRVFSLDTGRLNPETY+ FD VEKHYGI IEYMFPDAVEVQ
Sbjct: 378 FSGAEDVALIEYAKLTGRPFRVFSLDTGRLNPETYQLFDAVEKHYGIRIEYMFPDAVEVQ 557
Query: 175 GLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQVD 234
LVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSE+PVVQVD
Sbjct: 558 ALVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVD 737
Query: 235 PVFE 238
PVFE
Sbjct: 738 PVFE 749
>BE058404 similar to GP|18252504|gb adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (28%)
Length = 433
Score = 256 bits (655), Expect = 1e-68
Identities = 118/143 (82%), Positives = 131/143 (91%), Gaps = 3/143 (2%)
Frame = +1
Query: 251 NPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWEDAKA 310
+PVANVKGHDIW+FLRTMNVPVNSLH+KGY+SIGCEPCTR VLPGQHEREGRWWWEDAKA
Sbjct: 4 SPVANVKGHDIWNFLRTMNVPVNSLHAKGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 183
Query: 311 KECGLHKGNVKQDAEAELNGNGV--ANTNGTA-TVADIFNTQNVVSLSRTGIENLTKLEN 367
KECGLHKGNVKQ E ++NGNG+ ++ NG A TV DIFN+ NVV+LSRTGIENL +LE+
Sbjct: 184 KECGLHKGNVKQQKEEDVNGNGLSQSHANGDATTVPDIFNSPNVVNLSRTGIENLAELED 363
Query: 368 RKEPWLVVLYAPWCPYCQAMEES 390
RKEPWLVVLYAPWCPYCQAMEES
Sbjct: 364 RKEPWLVVLYAPWCPYCQAMEES 432
>TC219701 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate
reductase, partial (28%)
Length = 883
Score = 218 bits (556), Expect = 3e-57
Identities = 108/133 (81%), Positives = 115/133 (86%)
Frame = +1
Query: 329 NGNGVANTNGTATVADIFNTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAME 388
N N NG ATVADIF +QNVVSLSR+GIENL KLENRKE WLVVLYAPWC +CQAME
Sbjct: 1 NXNXXXQXNGXATVADIFISQNVVSLSRSGIENLAKLENRKEHWLVVLYAPWCRFCQAME 180
Query: 389 ESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENR 448
ESYVDLA+KLA SGVKV KFRADGEQKE+AK EL LGSFPTI+ FPKHSS+PIKYPSE R
Sbjct: 181 ESYVDLAEKLARSGVKVAKFRADGEQKEYAKSELQLGSFPTILLFPKHSSQPIKYPSEKR 360
Query: 449 DVDSLMAFVNALR 461
DVDSL AFVNALR
Sbjct: 361 DVDSLTAFVNALR 399
>TC221885 similar to UP|Q6V3B0 (Q6V3B0) Adenosine 5' phosphosulfate
reductase, partial (26%)
Length = 805
Score = 182 bits (462), Expect(2) = 4e-52
Identities = 92/113 (81%), Positives = 99/113 (87%), Gaps = 2/113 (1%)
Frame = +3
Query: 330 GNGVANTNGTAT-VADIFNTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAME 388
G ++ NG AT V DIFN+ NVV+LSRTGIENL KL +RKEPWLVVLYAPWCPYCQAME
Sbjct: 30 GLSXSHANGDATTVXDIFNSPNVVNLSRTGIENLAKLXDRKEPWLVVLYAPWCPYCQAME 209
Query: 389 ESYVDLADKLAES-GVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRP 440
ESYVDLADKLA S G+KVGKFRADGEQKEFAK EL LGSFPTI+FFPKHSSRP
Sbjct: 210 ESYVDLADKLAGSTGMKVGKFRADGEQKEFAKSELQLGSFPTILFFPKHSSRP 368
Score = 41.2 bits (95), Expect(2) = 4e-52
Identities = 19/20 (95%), Positives = 19/20 (95%)
Frame = +1
Query: 442 KYPSENRDVDSLMAFVNALR 461
KYPSE RDVDSLMAFVNALR
Sbjct: 376 KYPSEKRDVDSLMAFVNALR 435
>BG047004 similar to GP|18252504|gb adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (35%)
Length = 505
Score = 152 bits (384), Expect = 3e-37
Identities = 94/170 (55%), Positives = 116/170 (67%), Gaps = 11/170 (6%)
Frame = +2
Query: 2 ALAVTCSISISSSSSSSSTF-----QSSQPKFSQIASIRVSE--IPHGGGVNASQRRSFV 54
ALA + + S S+++++S +F SS K +I S R E + G VN +QR S V
Sbjct: 2 ALAASTTSSSSATAAASISFFSRLGSSSDAKAPKIGSFRFPERSLVSSGVVNVTQRHSLV 181
Query: 55 KP----PQRSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNH 110
+P PQR DSI PLAATIVA++V E E+++ +Q+ DLE AS L+IMD ALEKF N
Sbjct: 182 RPLNA*PQRI-DSIVPLAATIVATEV-ENEEEDVKQIVEDLEYASTLEIMDRALEKFEND 355
Query: 111 IAIAFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYG 160
IAIAFSGAEDVA IEYA LTGRP+RV SLDTGRLNPE + D EKHYG
Sbjct: 356 IAIAFSGAEDVAWIEYAHLTGRPYRVLSLDTGRLNPENLQIXDAGEKHYG 505
>TC210297 homologue to UP|Q8W1A1 (Q8W1A1) Adenosine 5'-phosphosulfate
reductase, partial (29%)
Length = 453
Score = 121 bits (304), Expect = 6e-28
Identities = 76/136 (55%), Positives = 98/136 (71%), Gaps = 11/136 (8%)
Frame = +2
Query: 1 MALAVTCSISISSSSSSSSTF-----QSSQPKFSQIASIRVSEIPH--GGGVNASQRRSF 53
+A++ T S S ++++++SS+F SS K QI S R E P G VN +QRRS
Sbjct: 50 LAVSTTSSSSAAAAAAASSSFFSRLGSSSDAKAPQIGSFRFPERPQVSSGVVNLTQRRSS 229
Query: 54 VKP----PQRSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGN 109
V+P PQR+ DS+ PLAATIVA +V E E+++++QLA DLEN+SPL+IMD ALEKFGN
Sbjct: 230 VRPLNAEPQRN-DSVVPLAATIVAPEV-EKEKEDFEQLAKDLENSSPLEIMDKALEKFGN 403
Query: 110 HIAIAFSGAEDVALIE 125
IAIAFSGAEDVALIE
Sbjct: 404 DIAIAFSGAEDVALIE 451
>BE555252
Length = 250
Score = 113 bits (282), Expect = 2e-25
Identities = 53/82 (64%), Positives = 64/82 (77%)
Frame = +2
Query: 330 GNGVANTNGTATVADIFNTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEE 389
GNG + NG+ATVAD FN+ +VV L+R+GIENL LEN +EPWLVVLYAPWC +C AM+E
Sbjct: 2 GNGASQANGSATVADFFNSHDVVRLTRSGIENLA*LENPQEPWLVVLYAPWCRFCHAMDE 181
Query: 390 SYVDLADKLAESGVKVGKFRAD 411
SYVDL+ + A SGV V F AD
Sbjct: 182 SYVDLSHQFAWSGVLVASFLAD 247
>TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like protein,
complete
Length = 1703
Score = 57.4 bits (137), Expect = 1e-08
Identities = 39/113 (34%), Positives = 55/113 (48%), Gaps = 1/113 (0%)
Frame = +3
Query: 347 NTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAES-GVKV 405
+ +VV LS +N K + LV YAPWC +C+ + Y L ++ V +
Sbjct: 189 SADDVVVLSE---DNFEKEVGQDRGALVEFYAPWCGHCKKLAPEYEKLGSSFKKAKSVLI 359
Query: 406 GKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVN 458
GK D E K + G+ +PTI +FPK S P KY R +SL+ FVN
Sbjct: 360 GKVDCD-EHKSLCSK-YGVSGYPTIQWFPKGSLEPKKYEGP-RTAESLVEFVN 509
Score = 53.9 bits (128), Expect = 1e-07
Identities = 39/125 (31%), Positives = 61/125 (48%), Gaps = 1/125 (0%)
Frame = +3
Query: 335 NTNGTATVADIFNTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDL 394
NT G V NVV L+ + L+ K+ LV YAPWC +C+++ +Y +
Sbjct: 507 NTEGGTNVKIATVPSNVVVLTPENFNEVV-LDETKDV-LVEFYAPWCGHCKSLAPTYEKV 680
Query: 395 ADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSL 453
A E V + AD + ++ A++ + FPT+ FFPK + Y RD+D
Sbjct: 681 ATAFKLEEDVVIANLDAD-KYRDLAEK-YDVSGFPTLKFFPKGNKAGEDY-GGGRDLDDF 851
Query: 454 MAFVN 458
+AF+N
Sbjct: 852 VAFIN 866
>AI930908 similar to GP|18252504|gb| adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (16%)
Length = 404
Score = 55.8 bits (133), Expect(2) = 1e-08
Identities = 33/64 (51%), Positives = 40/64 (61%)
Frame = +1
Query: 248 VKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVLPGQHEREGRWWWED 307
VKWNPVANV FLRTM+VPV + Y+ + EPCT + GQHER+ WWED
Sbjct: 79 VKWNPVANVMV*TYGVFLRTMDVPVIIAFPR-YV*L-LEPCT-SQFTGQHERKEGVWWED 249
Query: 308 AKAK 311
AK +
Sbjct: 250 AKPR 261
Score = 43.9 bits (102), Expect = 2e-04
Identities = 39/118 (33%), Positives = 54/118 (45%), Gaps = 6/118 (5%)
Frame = +2
Query: 229 PVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDI------WSFLRTMNVPVNSLHSKGYIS 282
P VDPVFEG+DGG+GSL + + + W FL LHS+
Sbjct: 23 PCCPVDPVFEGLDGGIGSL*SGTRLQMLWSRHMESSLGPWMFL*L-------LHSQDMFD 181
Query: 283 IGCEPCTRAVLPGQHEREGRWWWEDAKAKECGLHKGNVKQDAEAELNGNGVANTNGTA 340
C +A LP + +E + +ECGLHKGN+K + NG +NG+A
Sbjct: 182 --CWSRAQASLP-VNMKERKVCGGRMPNQECGLHKGNIKHEICHLWNG---PPSNGSA 337
Score = 21.2 bits (43), Expect(2) = 1e-08
Identities = 8/10 (80%), Positives = 10/10 (100%)
Frame = +3
Query: 224 TRSEVPVVQV 233
TRSE+PVVQ+
Sbjct: 9 TRSEIPVVQL 38
>TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like protein,
complete
Length = 1422
Score = 54.3 bits (129), Expect = 1e-07
Identities = 36/113 (31%), Positives = 53/113 (46%), Gaps = 1/113 (0%)
Frame = +1
Query: 347 NTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAES-GVKV 405
+ +VV+L+ EN + LV YAPWC +C+ + Y L ++ V +
Sbjct: 187 SADDVVALTEETFENEV---GKDRAALVEFYAPWCGHCKRLAPEYEQLGASFKKTKSVLI 357
Query: 406 GKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVN 458
K D + K G+ +PTI +FPK S P KY R ++L AFVN
Sbjct: 358 AKVDCDEHKSVCGK--YGVSGYPTIQWFPKGSLEPKKYEGA-RTAEALAAFVN 507
Score = 52.4 bits (124), Expect = 4e-07
Identities = 30/87 (34%), Positives = 47/87 (53%), Gaps = 1/87 (1%)
Frame = +1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
LV YAPWC +C+A+ Y +A + V + AD + K+ A++ G+ +PT+
Sbjct: 613 LVEFYAPWCGHCKALAPIYEKVAAAFNLDKDVVIANVDAD-KYKDLAEK-YGVSGYPTLK 786
Query: 432 FFPKHSSRPIKYPSENRDVDSLMAFVN 458
FFPK + Y RD+D +AF+N
Sbjct: 787 FFPKSNKAGENYDG-GRDLDDFVAFIN 864
>BG362767
Length = 491
Score = 50.8 bits (120), Expect = 1e-06
Identities = 29/86 (33%), Positives = 46/86 (52%), Gaps = 1/86 (1%)
Frame = +2
Query: 374 VVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
V YAPWC +C+A+ Y +A + V + AD + K+ A++ G+ +PT+ F
Sbjct: 59 VEFYAPWCGHCKALAPIYEKVAAAFNLDKDVVIANVDAD-KYKDLAEK-YGVSGYPTLKF 232
Query: 433 FPKHSSRPIKYPSENRDVDSLMAFVN 458
FPK + Y RD+D +AF+N
Sbjct: 233 FPKSNKAGENYDG-GRDLDDFVAFIN 307
>TC206923 similar to UP|Q8LAM5 (Q8LAM5) Protein disulfide isomerase-like,
partial (53%)
Length = 1422
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/80 (32%), Positives = 36/80 (44%)
Frame = +1
Query: 361 NLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKR 420
N T + +V YAPWC +CQA+ Y A +L GV + K A E +
Sbjct: 445 NFTTVVENNRFIMVEFYAPWCGHCQALAPEYAAAATELKPDGVVLAKVDATVENE--LAN 618
Query: 421 ELGLGSFPTIMFFPKHSSRP 440
E + FPT+ FF +P
Sbjct: 619 EYDVQGFPTVFFFVDGVHKP 678
>TC207250 similar to UP|Q9LW75 (Q9LW75) Dbj|BAA81871.1, partial (26%)
Length = 927
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/81 (30%), Positives = 40/81 (48%)
Frame = +1
Query: 360 ENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAK 419
EN ++ N E LV+ YAPWCP + + + A L E G + + D ++
Sbjct: 436 ENTERVVNGNEFVLVLGYAPWCPRSAELMPHFAEAATSLKELGSPLIMAKLDADRYPKPA 615
Query: 420 RELGLGSFPTIMFFPKHSSRP 440
LG+ FPT++ F +S+P
Sbjct: 616 SFLGVKGFPTLLLFVNGTSQP 678
>TC207075 similar to UP|Q84XU4 (Q84XU4) Protein disulphide isomerase
(Fragment), partial (30%)
Length = 928
Score = 46.2 bits (108), Expect = 3e-05
Identities = 27/72 (37%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Frame = +3
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAE-SGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQA+E +Y LA L + + K + AK + FPT++
Sbjct: 96 LLEIYAPWCGHCQALEPTYNKLAKHLRSIESIVIAKMDGTTNEHPRAKSD----GFPTLL 263
Query: 432 FFP--KHSSRPI 441
FFP SS PI
Sbjct: 264 FFPAGNKSSDPI 299
>TC218708 similar to GB|AAF40463.1|7211992|F13M7 Strong simialrity to the
disulfide isomerase precursor homolog T21L14.14
gi|2702281 from A. thaliana on BAC gb|AC003033.
{Arabidopsis thaliana;} , partial (77%)
Length = 1274
Score = 45.1 bits (105), Expect = 7e-05
Identities = 23/75 (30%), Positives = 38/75 (50%)
Frame = +2
Query: 369 KEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFP 428
KE W+V +APWC +C+ + + ++ L + VK+G D E+ ++ + FP
Sbjct: 275 KELWIVEFFAPWCGHCKKLAPEWKKASNNL-KGKVKLGHVDCDAEKSLMSR--FKVQGFP 445
Query: 429 TIMFFPKHSSRPIKY 443
TI+ F PI Y
Sbjct: 446 TILVFGADKDSPIPY 490
>TC216352 protein disulfide isomerase-like protein [Glycine max]
Length = 2032
Score = 44.3 bits (103), Expect = 1e-04
Identities = 27/85 (31%), Positives = 43/85 (49%)
Frame = +1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
+V YAPWC +CQA+ Y A +L V + K A E+ E A ++ + FPT+ F
Sbjct: 409 MVEFYAPWCGHCQALAPEYAAAATELKGEDVILAKVDAT-EENELA-QQYDVQGFPTVHF 582
Query: 433 FPKHSSRPIKYPSENRDVDSLMAFV 457
F +P + R D++M ++
Sbjct: 583 FVDGIHKPY---NGQRTKDAIMTWI 648
Score = 41.6 bits (96), Expect = 8e-04
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 1/63 (1%)
Frame = +1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAE-SGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQ++E Y LA L + + K + AK + FPT++
Sbjct: 1426 LLEIYAPWCGHCQSLEPIYNKLAKHLRNIDSLVIAKMDGTTNEHPRAKPD----GFPTLL 1593
Query: 432 FFP 434
FFP
Sbjct: 1594 FFP 1602
>TC229461 similar to UP|Q84XU4 (Q84XU4) Protein disulphide isomerase
(Fragment), partial (29%)
Length = 766
Score = 42.0 bits (97), Expect = 6e-04
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 1/63 (1%)
Frame = +1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAE-SGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQ++E Y LA L + + K + AK + FPT++
Sbjct: 70 LLXIYAPWCGHCQSLEPIYNKLAKHLRNIDSLVIAKMDGTTNEHPRAKPD----GFPTLL 237
Query: 432 FFP 434
FFP
Sbjct: 238 FFP 246
>TC207907 similar to UP|Q96313 (Q96313) AT.I.24-13 protein, partial (80%)
Length = 858
Score = 38.9 bits (89), Expect = 0.005
Identities = 23/95 (24%), Positives = 44/95 (46%), Gaps = 1/95 (1%)
Frame = +2
Query: 364 KLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKREL 422
K++ + W V PWC +C+ + + DL + E ++VG+ ++ +K +
Sbjct: 191 KIKEKDTAWFVKFCVPWCKHCKNLGSLWDDLGKAMEGEDEIEVGEVDCGMDKAVCSK--V 364
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
+ S+PT F + + RDV+S+ FV
Sbjct: 365 DIHSYPTFKVF--YDGEEVARYQGTRDVESMKTFV 463
>BI969216 weakly similar to GP|28140231|gb protein disulphide isomerase
{Elaeis guineensis}, partial (26%)
Length = 634
Score = 37.4 bits (85), Expect = 0.014
Identities = 21/62 (33%), Positives = 30/62 (47%)
Frame = -1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
L+ +Y PWC +CQ+ E LA+ V + DG E + + L FPT+ F
Sbjct: 571 LLEIYVPWCXHCQSXEPISTKLANXXXNIDFLV-IAKMDGPPNELPR--VKLDGFPTLFF 401
Query: 433 FP 434
FP
Sbjct: 400 FP 395
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,658,581
Number of Sequences: 63676
Number of extensions: 272457
Number of successful extensions: 2853
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 2303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2578
length of query: 461
length of database: 12,639,632
effective HSP length: 101
effective length of query: 360
effective length of database: 6,208,356
effective search space: 2235008160
effective search space used: 2235008160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0026.11


