

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
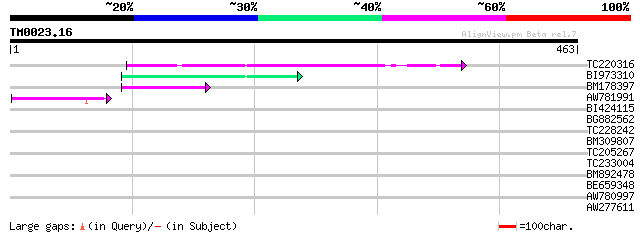
Query= TM0023.16
(463 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220316 92 4e-19
BI973310 56 3e-08
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 44 1e-04
AW781991 44 2e-04
BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidop... 40 0.002
BG882562 37 0.014
TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired r... 35 0.054
BM309807 similar to GP|5764395|gb| far-red impaired response pro... 35 0.054
TC205267 similar to GB|AAM10267.1|20147101|AY091668 AT4g38690/F2... 33 0.35
TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] ... 32 0.60
BM892478 31 1.3
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 29 3.9
AW780997 29 5.1
AW277611 28 6.6
>TC220316
Length = 988
Score = 92.4 bits (228), Expect = 4e-19
Identities = 68/278 (24%), Positives = 126/278 (44%)
Frame = +1
Query: 96 WDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRY 155
W+ GL+D+ W+KE Y+KR+ WV +L+G FFAG E S G ++++
Sbjct: 46 WNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGIPMN---ESLDSFFGALLNAQT 216
Query: 156 NLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPV 215
L +F+ + + ++ R E ++D ++ + P ++T E +E + +T VF +F+
Sbjct: 217 PLMEFIPRYERGLERRREEERKEDFNTSNFQPILQTK-EPVEEQCRRLYTLTVFKIFQKE 393
Query: 216 LQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEH 275
L + Y V + ++ V+ ++ N SC E G+ C H
Sbjct: 394 LLQCFSYLGFKIFEEGGLSRYMVRRCGNDMEKHVVTFNASNISISCSCQMFEYEGVLCRH 573
Query: 276 IVDVLAHLRIEELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALG 335
++ V L++ E+ +I RW++ +DGV+ + + W S+ +L +
Sbjct: 574 VLRVFQILQLREVPSRYILHRWTRNTEDGVFP----DMESW--------SSSQELKNLML 717
Query: 336 YLNSNTAADFNDATQKASELIEQSKAKKSCQREGSLSL 373
+ TA+ + DA + IE+ K REG L
Sbjct: 718 WSLRETASKYIDA---GATSIEKYKLAYEILREGGRKL 822
>BI973310
Length = 457
Score = 56.2 bits (134), Expect = 3e-08
Identities = 37/148 (25%), Positives = 59/148 (39%)
Frame = +3
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F W L+ + DN W++ Y R WV +LR FF E S +V
Sbjct: 15 FESYWHSLLERFYVMDNEWLQSIYNSRQHWVPVYLRETFFGEISLNEGNEYLISFFDGYV 194
Query: 152 HSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLL 211
S L ++Q + V +E++ D + + P +KT +E A +T +F+
Sbjct: 195 FSSTTLQVLVRQYEKAVSSWHEKELKADYDTTNSSPVLKTP-SPMEKQAASLYTRKIFMK 371
Query: 212 FRPVLQHAPLMKVLDCTDAVTCLNYTVA 239
F+ L + D+ T Y VA
Sbjct: 372 FQEELVETLANPAIKIDDSGTITTYRVA 455
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/73 (27%), Positives = 35/73 (47%)
Frame = +1
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F W E+ + GL N + Y RS+W LR F AG TT + + + + + +
Sbjct: 178 FELGWREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAGMTTTGQSKSINAFIQRFL 357
Query: 152 HSRYNLTDFLQQL 164
++ L F++Q+
Sbjct: 358 SAQTRLAHFVEQV 396
>AW781991
Length = 419
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/85 (24%), Positives = 44/85 (51%), Gaps = 3/85 (3%)
Frame = +3
Query: 2 LKNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
L+ D + + ++ ++ +++ N+F D+ S+ ++FW D R++Y +GD + FD T
Sbjct: 105 LEGDIQLVLDYLRQMHAENPNFFYAVQGDEDQSITNVFWADPKARMNYTFFGDTVTFDTT 284
Query: 62 ---EKYLCAWHLLRNANQNLGNPLL 83
+Y + N + G P+L
Sbjct: 285 YRSNRYRLPFAFFTGVNHH-GQPVL 356
>BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (9%)
Length = 402
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/68 (29%), Positives = 33/68 (48%)
Frame = +2
Query: 236 YTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFK 295
+ VAK K + V+++ SC E G+ C+HI+ V + L +I K
Sbjct: 14 FRVAKFEDDQKAYMVTLNHSELKANCSCQMFEYAGILCKHILTVFTVTNVLTLPPHYILK 193
Query: 296 RWSKGAKD 303
RW++ AK+
Sbjct: 194 RWTRNAKN 217
>BG882562
Length = 416
Score = 37.4 bits (85), Expect = 0.014
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = +3
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
K D + + + + N+F + L+++FW D R +Y +GDV+AFD T
Sbjct: 24 KGDPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDST 200
>TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein; Mutator-like transposase-like protein;
phytochrome A signaling protein-like, partial (25%)
Length = 1354
Score = 35.4 bits (80), Expect = 0.054
Identities = 20/68 (29%), Positives = 30/68 (43%)
Frame = +2
Query: 246 KEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAKDGV 305
K++ V V+ + C E RG C H + VL + +I KRW+K AK
Sbjct: 197 KDFFVVVNQVKSELSCICRLFEYRGYLCRHALFVLQYSGQSVFPSQYILKRWTKDAKVRN 376
Query: 306 YSGKNIEN 313
G+ E+
Sbjct: 377 IMGEESEH 400
>BM309807 similar to GP|5764395|gb| far-red impaired response protein
{Arabidopsis thaliana}, partial (15%)
Length = 428
Score = 35.4 bits (80), Expect = 0.054
Identities = 30/127 (23%), Positives = 54/127 (41%), Gaps = 7/127 (5%)
Frame = +3
Query: 183 IHGVPAMKTNLESL-EMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLNYTVAKL 241
IH + NL L E + +T+ +F F+ ++ V C + + T+AK
Sbjct: 3 IHCTNSQHLNLLHLXEKQMSTVYTHAIFKKFQ-----VEVLGVAGCQSRIEAGDGTIAK- 164
Query: 242 SFLAKEWH------VSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFK 295
F+ +++ V+ + + + C E +G C H + VL + +I K
Sbjct: 165 -FIVQDYEKDEEFLVTWNELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSCVPSHYILK 341
Query: 296 RWSKGAK 302
RW+K AK
Sbjct: 342 RWTKDAK 362
>TC205267 similar to GB|AAM10267.1|20147101|AY091668 AT4g38690/F20M13_250
{Arabidopsis thaliana;} , partial (92%)
Length = 1403
Score = 32.7 bits (73), Expect = 0.35
Identities = 17/81 (20%), Positives = 40/81 (48%)
Frame = +3
Query: 7 VMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEKYLC 66
++ ++ E +D F +++ +Q+ +HL +D V+G +A + ++ +C
Sbjct: 576 ILILEIRTEFGHEDPPEFDKYLEEQLG--EHLIHQDDA------VFGKTVAELLPKRVIC 731
Query: 67 AWHLLRNANQNLGNPLLTKGF 87
W +++ G+PL + GF
Sbjct: 732 VWKPRKSSQPKPGSPLWSAGF 794
>TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 551
Score = 32.0 bits (71), Expect = 0.60
Identities = 29/160 (18%), Positives = 61/160 (38%), Gaps = 7/160 (4%)
Frame = +1
Query: 150 HVHSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVF 209
+VH + +F+ + + +E D + + +KT + E+ K +T +F
Sbjct: 7 YVHKHTSFKEFVDKYDLVLHRKHLKEAMADLETRNVSFELKTRC-NFEVQLAKVYTKEIF 183
Query: 210 LLFRPVLQHA-PLMKVLDCTDAVTCLNYTVAKLSFL------AKEWHVSVSSDNKDFKYS 262
F+ ++ + + + Y V + + K + V + D +
Sbjct: 184 QKFQSEVEGMYSCFNTRQVSVNGSIITYVVKERVEVEGNEKGVKSFEVLYETTELDIRCI 363
Query: 263 CMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAK 302
C +G C ++VL + IEE+ +I RW + K
Sbjct: 364 CSLFNYKGYLCRRALNVLNYNGIEEIPSRYILHRWRRDFK 483
>BM892478
Length = 429
Score = 30.8 bits (68), Expect = 1.3
Identities = 15/52 (28%), Positives = 26/52 (49%)
Frame = +3
Query: 10 MQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
+++ ++D +F D N + ++FW DG R S +GDVL D +
Sbjct: 33 LEYFQSRQAEDTGFFYAMEVDNGNCM-NIFWADGRSRYSCSHFGDVLVLDTS 185
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 29.3 bits (64), Expect = 3.9
Identities = 13/36 (36%), Positives = 16/36 (44%)
Frame = -1
Query: 167 CVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATK 202
C R +D C+SIHG P N+ E S K
Sbjct: 383 CSYCKRVGHTQDTCYSIHGFPGKSVNISKSETSEIK 276
>AW780997
Length = 422
Score = 28.9 bits (63), Expect = 5.1
Identities = 19/62 (30%), Positives = 26/62 (41%), Gaps = 9/62 (14%)
Frame = +2
Query: 222 MKVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYS---------CMKMESRGLA 272
+ +LD V C T+ K W+V SDN DFK C KM +R +
Sbjct: 179 VNLLDLRGTVLC---TIHKKLLAFGRWNVYRGSDNSDFKIQEXAGFQVKRCYKMITRKVT 349
Query: 273 CE 274
C+
Sbjct: 350 CQ 355
>AW277611
Length = 442
Score = 28.5 bits (62), Expect = 6.6
Identities = 16/41 (39%), Positives = 20/41 (48%)
Frame = +1
Query: 383 DPIRLRRKGRGTKNKSSLGLTVKHVINCLIFKTPGHNKLTG 423
D +R R RGT+ KS + +HV L PGH L G
Sbjct: 262 DYVRRGRNSRGTEQKS*VRTASQHVQRQLREPDPGHEHLQG 384
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,689,377
Number of Sequences: 63676
Number of extensions: 341868
Number of successful extensions: 1448
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1446
length of query: 463
length of database: 12,639,632
effective HSP length: 101
effective length of query: 362
effective length of database: 6,208,356
effective search space: 2247424872
effective search space used: 2247424872
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0023.16


