

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.15
(528 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
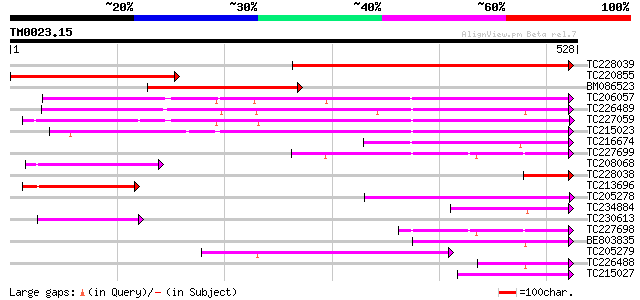
Score E
Sequences producing significant alignments: (bits) Value
TC228039 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {A... 483 e-137
TC220855 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {A... 296 1e-80
BM086523 253 1e-67
TC206057 homologue to UP|Q9ZRX1 (Q9ZRX1) Cytosolic chaperonin, d... 246 2e-65
TC226489 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1... 233 2e-61
TC227059 similar to UP|O81503 (O81503) F9D12.18 protein, partial... 219 2e-57
TC215023 similar to UP|Q6PBW6 (Q6PBW6) Chaperonin containing TCP... 197 7e-51
TC216674 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/... 96 4e-20
TC227699 similar to UP|TCPZ_HUMAN (P40227) T-complex protein 1, ... 94 1e-19
TC208068 homologue to PRF|2206327A.0|1587206|2206327A T complex ... 94 1e-19
TC228038 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {A... 87 1e-17
TC213696 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/... 84 2e-16
TC205278 similar to PRF|2206327A.0|1587206|2206327A T complex pr... 81 1e-15
TC234884 similar to UP|TCPA_DROME (P12613) T-complex protein 1, ... 77 1e-14
TC230613 homologue to UP|Q8L7N0 (Q8L7N0) TCP-1 chaperonin-like p... 74 1e-13
TC227698 similar to UP|TCPZ_HUMAN (P40227) T-complex protein 1, ... 72 6e-13
BE803835 similar to GP|24371053|db t-complex polypeptide 1 {Brug... 71 1e-12
TC205279 homologue to PRF|2206327A.0|1587206|2206327A T complex ... 68 1e-11
TC226488 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1... 61 1e-09
TC215027 weakly similar to UP|Q8WSY6 (Q8WSY6) CCT chaperonin bet... 60 3e-09
>TC228039 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {Arabidopsis
thaliana;} , partial (50%)
Length = 1141
Score = 483 bits (1244), Expect = e-137
Identities = 245/262 (93%), Positives = 254/262 (96%)
Frame = +2
Query: 264 AEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFEL 323
AEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFEL
Sbjct: 2 AEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFEL 181
Query: 324 RRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGST 383
RRFCRTTG+VA+LKL QPNPDDLGYVDSVSV+E+GGV VTIVKNEEGGNSV+TVVLRGST
Sbjct: 182 RRFCRTTGSVAMLKLGQPNPDDLGYVDSVSVQEIGGVRVTIVKNEEGGNSVATVVLRGST 361
Query: 384 DSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAIAKF 443
DSILDDLERAVDDGVNTYKAMC+DSR VPGAAATEIELAKRVKDFSFKETGLDQYAIAKF
Sbjct: 362 DSILDDLERAVDDGVNTYKAMCRDSRFVPGAAATEIELAKRVKDFSFKETGLDQYAIAKF 541
Query: 444 AESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVT 503
AESFEMIPRTLAENAGLNAMEIISSLYAEHASGN KVGIDLEEGVCKDV+T +WDLHVT
Sbjct: 542 AESFEMIPRTLAENAGLNAMEIISSLYAEHASGNAKVGIDLEEGVCKDVSTLSIWDLHVT 721
Query: 504 KFFALKYAADAVCTVLRVDQVI 525
K FALKYAADA CTVLRVDQ+I
Sbjct: 722 KLFALKYAADAACTVLRVDQII 787
>TC220855 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {Arabidopsis
thaliana;} , partial (28%)
Length = 575
Score = 296 bits (759), Expect = 1e-80
Identities = 149/158 (94%), Positives = 154/158 (97%)
Frame = +2
Query: 1 MGFNMQSYGIQSMLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLD 60
MGFN+Q YGIQSMLKEGHKHLSGLDEAV+KNIDACKQLSTITRTSLGPNGMNKMVINHLD
Sbjct: 89 MGFNLQPYGIQSMLKEGHKHLSGLDEAVLKNIDACKQLSTITRTSLGPNGMNKMVINHLD 268
Query: 61 KLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRM 120
KLFVTNDA TIVNELEVQHPAAK+LVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRM
Sbjct: 269 KLFVTNDAGTIVNELEVQHPAAKVLVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRM 448
Query: 121 GLHPSEIISGYTKAIAKTIEILDELVEKGSESMDVRDK 158
GLHPSEIISGYTKAI KT++ILDELVE GSESMDVRDK
Sbjct: 449 GLHPSEIISGYTKAINKTVQILDELVENGSESMDVRDK 562
>BM086523
Length = 433
Score = 253 bits (647), Expect = 1e-67
Identities = 128/144 (88%), Positives = 138/144 (94%)
Frame = +2
Query: 129 SGYTKAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADAC 188
SGYTKAI KT++ILDELVE GS++MDVRDK QVISRM+AAVASKQFGQED ICSLVADAC
Sbjct: 2 SGYTKAINKTVQILDELVEDGSDNMDVRDKEQVISRMKAAVASKQFGQEDIICSLVADAC 181
Query: 189 IQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFVSGV 248
IQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRG+VLKSDAVG+IK+ EKAKVAVF SGV
Sbjct: 182 IQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGLVLKSDAVGTIKQAEKAKVAVFASGV 361
Query: 249 DTSATDTKGTVLIHSAEQLENYSK 272
DTSAT+TKGTVLIH+AEQLENYSK
Sbjct: 362 DTSATETKGTVLIHTAEQLENYSK 433
>TC206057 homologue to UP|Q9ZRX1 (Q9ZRX1) Cytosolic chaperonin,
delta-subunit, complete
Length = 1930
Score = 246 bits (628), Expect = 2e-65
Identities = 150/507 (29%), Positives = 271/507 (52%), Gaps = 12/507 (2%)
Frame = +2
Query: 31 NIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGK 90
NI A + ++ RTSLGP GM+KM+ D++ +TND ATI+N+++V PAAK+LV K
Sbjct: 170 NIVAARSVANAVRTSLGPKGMDKMISTSSDEVIITNDGATILNKMQVLQPAAKMLVELSK 349
Query: 91 AQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKGS 150
+Q GDG + AG LL+ L+ G+HP+ + KA K +++L + +
Sbjct: 350 SQDSAAGDGTTTVVVIAGALLEQCLLLLSHGIHPTVVSDALHKAAVKAVDVLTAM----A 517
Query: 151 ESMDVRDKVQVISRMRAAVASKQFGQEDTICS-LVADACIQV--CPKNPANFNVDNVRVA 207
+++ D+ ++ ++ SK Q T+ + L DA + V PK P ++ +V++
Sbjct: 518 VPVELSDRDSLVKSASTSLNSKVVSQYSTLLAPLAVDAVLSVVDAPK-PDMVDLRDVKIV 694
Query: 208 KLLGGGLHNSTVVRGMVLK---SDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHSA 264
K LGG + ++ +V+G+V S A G R+E AK+AV + TD + ++++
Sbjct: 695 KKLGGTVDDTELVKGLVFDKKVSHAAGGPTRMENAKIAVIQFQISPPKTDIEQSIVVSDY 874
Query: 265 EQLENYSKTEEAKVEELIKAVADSGAKVI-----VSGGAVGEMALHFCERYKLMVLKISS 319
Q++ K E + + +IK + +G V+ + AV +++LH+ + K++V+K
Sbjct: 875 SQMDRILKEERSYILSMIKKIKATGCNVLLIQKSILRDAVTDLSLHYLAKAKILVIKDVE 1054
Query: 320 KFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVG-GVTVTIVKNEEGGNSVSTVV 378
+ E+ +T + + + + LGY D V +G G V I +E G + +TV+
Sbjct: 1055RDEIEFITKTLNCLPIANIEHFRTEKLGYADLVEEFSLGDGKIVKITGIKEMGKT-TTVL 1231
Query: 379 LRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQY 438
+RGS +LD+ ER++ D + + + + G A EIEL++++ ++ G++ Y
Sbjct: 1232VRGSNQLVLDEAERSLHDALCVVRCLVAKRFLIAGGGAPEIELSRQLGAWAKVLHGMEGY 1411
Query: 439 AIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVW 498
+ FAE+ E+IP TLAENAGLN + I++ L HA G GI++ +G ++ +V
Sbjct: 1412CVRAFAEALEVIPYTLAENAGLNPIAIVTELRNRHAQGEINAGINVRKGQITNILEENVV 1591
Query: 499 DLHVTKFFALKYAADAVCTVLRVDQVI 525
+ A+ A + V +L++D ++
Sbjct: 1592QPLLVSTSAITLATECVRMILKIDDIV 1672
>TC226489 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1, complete
Length = 2038
Score = 233 bits (594), Expect = 2e-61
Identities = 152/516 (29%), Positives = 269/516 (51%), Gaps = 20/516 (3%)
Frame = +1
Query: 30 KNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAG 89
+N+ AC+ ++ I ++SLGP G++KM+++ + + +TND ATI+ LEV+HPAAK+LV
Sbjct: 136 QNVVACQAVANIVKSSLGPVGLDKMLVDDIGDVTITNDGATILKMLEVEHPAAKVLVELA 315
Query: 90 KAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKG 149
+ Q E+GDG + A ELL+ A +L+R +HP+ IISGY A+ + + ++ EK
Sbjct: 316 ELQDREVGDGTTSVVIVAAELLKRANDLVRNKIHPTSIISGYRLAMREACKYVE---EKL 486
Query: 150 SESMDVRDKVQVISRMRAAVASKQF-GQEDTICSLVADACIQVCPKN---PANFNVDNVR 205
+ ++ K +I+ + +++SK G D LV DA V N + + +
Sbjct: 487 AVKVEKLGKDSLINCAKTSMSSKLIAGDSDFFAILVVDAVQAVKMTNARGEVKYPIKGIN 666
Query: 206 VAKLLGGGLHNSTVVRGMVLKSD--AVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHS 263
+ K G +S ++ G L + A G R+ A++A + + VL+
Sbjct: 667 ILKAHGKSARDSFLMNGYALNTGRAAQGMPLRVAPARIACLDFNLQKTKMQLGVQVLVTD 846
Query: 264 AEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFEL 323
+LE + E +E I+ + +GA VI++ + +MAL + + ++ K ++
Sbjct: 847 PRELEKIRQREADMTKERIEKLLKAGANVILTTKGIDDMALKYFVEAGAIAVRRVRKEDM 1026
Query: 324 RRFCRTTGAVAVLKLSQP------NPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTV 377
R + TGA V + P LGY D V E + V ++K + ++V T+
Sbjct: 1027RHVAKATGATLVSTFADMEGEETFEPSFLGYADEVVEERISDDAVVMIKGTKTTSAV-TL 1203
Query: 378 VLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQ 437
+LRG+ D +LD+++RA+ D ++ K + + V G A E L+ ++ + +Q
Sbjct: 1204ILRGANDHMLDEMDRALHDALSIVKRTLESNTVVAGGGAVEAALSVYLEYLATTLGSREQ 1383
Query: 438 YAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTK--------VGIDLEEGVC 489
AIA+FAES +IP+ L+ NA +A E+++ L A H S TK +G+DL EG
Sbjct: 1384LAIAEFAESLLIIPKVLSVNAAKDATELVAKLRAYHHSAQTKADKKHLSSMGLDLSEGKI 1563
Query: 490 KDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
++ V + ++K +++A +A T+LR+D +I
Sbjct: 1564RNNLEAGVIEPAMSKVKIIQFATEAAITILRIDDMI 1671
>TC227059 similar to UP|O81503 (O81503) F9D12.18 protein, partial (92%)
Length = 2147
Score = 219 bits (558), Expect = 2e-57
Identities = 144/524 (27%), Positives = 275/524 (52%), Gaps = 10/524 (1%)
Frame = +2
Query: 13 MLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIV 72
+LK+ K SG + I A K ++ + RT+LGP M KM+++ + VTND I+
Sbjct: 218 VLKDSLKRESG-SKVRYAIIQAAKAVADVVRTTLGPRSMLKMLLDAQGGIVVTNDGNAIL 394
Query: 73 NELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYT 132
EL++ HPAAK ++ + Q EE+GDG I AGE+L A+ I +HP+ I Y
Sbjct: 395 RELDLAHPAAKSMIELSRTQDEEVGDGTTSVIILAGEMLHVADAFIDK-IHPTVICRAYA 571
Query: 133 KAIAKTIEILDELVEKGSESMDVRDKVQVISRMRAAVASKQFGQ-EDTICSLVADACIQV 191
KA+ I +LD++ + ++ +D+ ++ +++ + +K G+ D I L DA V
Sbjct: 572 KALEDAIAVLDKI----AMPINAQDRGIMLGLVKSCIGTKFTGRFGDLIADLAIDATTTV 739
Query: 192 ---CPKNPANFNVDN-VRVAKLLGGGLHNSTVVRGMVLKSDAV--GSIKR-IEKAKVAVF 244
+ + ++ N ++V K+ GG L +S V++G+++ D V G ++R I ++ +
Sbjct: 740 GVEVGQGLRDVDIKNYIKVEKVPGGQLEDSRVLKGVMINKDVVAPGKMRRKIVNPRIILL 919
Query: 245 VSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMAL 304
++ + + + E K EE +EEL + ++++ + ++A
Sbjct: 920 DCPLEYKKGENQTNAELLKEEDWSLLLKMEEEYIEELCMQILKFKPDLVITEKGLSDLAC 1099
Query: 305 HFCERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSV-SVEEVGGVTVT 363
H+ ++ + ++ K + R + GAV V + + D+G + V+++G
Sbjct: 1100HYLSKHGVSAIRRLRKTDNNRIAKACGAVIVNRPDELQESDVGTGAGLFEVKKIGDEYFA 1279
Query: 364 IVKNEEGGNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAK 423
+ + + + TV+LRG++ +L+++ER + D ++ + + K+ + VPG ATE+ ++
Sbjct: 1280FIVDCKEPKAC-TVLLRGASKDLLNEVERNLQDAMSVARNIIKNPKLVPGGGATELTVSA 1456
Query: 424 RVKDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASG-NTKVGI 482
+K S G++++ A +FE IPRTLA+N G+N + +++L +HA+G N +GI
Sbjct: 1457ALKQKSSSIEGIEKWPYEAAALAFEAIPRTLAQNCGVNVIRTMTALQGKHANGENAWIGI 1636
Query: 483 DLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVIN 526
D G D+ +WD + K A K A +A C +LR+D +++
Sbjct: 1637DGNTGSITDMKECKIWDAYNVKAQAFKTAIEAACMLLRIDDIVS 1768
>TC215023 similar to UP|Q6PBW6 (Q6PBW6) Chaperonin containing TCP1, subunit 2
(Beta), partial (98%)
Length = 1992
Score = 197 bits (502), Expect = 7e-51
Identities = 139/494 (28%), Positives = 240/494 (48%), Gaps = 6/494 (1%)
Frame = +3
Query: 38 LSTITRTSLGPNGMNKMV--INHLDKLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEE 95
++ + +T+LGP GM+K++ ++ VTND ATI+ L + +PAAK+LV K Q +E
Sbjct: 183 IADLVKTTLGPKGMDKILQSTGRGREVTVTNDGATILKSLHIDNPAAKVLVDISKVQDDE 362
Query: 96 IGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAI-AKTIEILDELVEKGSESMD 154
+GDG + AGELL+ AE+L+ +HP IISG+ A +L+++V+ ++S
Sbjct: 363 VGDGTTSVVVLAGELLREAEKLVATKIHPMTIISGFRMAAECARNALLEKVVDNKADSEK 542
Query: 155 VRDKVQVISRMRAAVASKQFGQE-DTICSLVADACIQVCPKNPANFNVDNVRVAKLLGGG 213
R + I+ ++SK Q+ + L DA +++ + N++++++ K GG
Sbjct: 543 FRSDLLNIAM--TTLSSKILSQDKEHFAKLAVDAVMRL----KGSTNLESIQIIKKPGGS 704
Query: 214 LHNSTVVRGMVL-KSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGT-VLIHSAEQLENYS 271
L +S + G +L K +G KRIE AK+ V + +DT G V + S ++
Sbjct: 705 LMDSFLDEGFILDKKIGIGQPKRIENAKILVANTAMDTDKVKIYGARVRVDSMARVAQIE 884
Query: 272 KTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFELRRFCRTTG 331
E+ K+ E ++ + G V+ + ++ ++ + + R TG
Sbjct: 885 TAEKEKMREKVQKIIGHGINCFVNRQLIYNFPEELFADAGILAIEHADFDGIERLALVTG 1064
Query: 332 AVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILDDLE 391
P LG+ D + +G + G + T+VLRG++ +LD+ E
Sbjct: 1065GEIASTFDNPESVKLGHCDLIEEIMIGEDKLIHFSGVAMGQAC-TIVLRGASHHVLDEAE 1241
Query: 392 RAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEMIP 451
R++ D + DSR + G E+ +AK V + K G AI F+ + IP
Sbjct: 1242RSLHDALCVLSQTVNDSRVLLGGGWPEMVMAKEVDALAKKTPGKKSLAIEAFSRALLAIP 1421
Query: 452 RTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYA 511
+A+NAGL++ E+IS L AEH GID+ G D+ + + K L +
Sbjct: 1422TIIADNAGLDSAELISQLRAEHQKEGCTAGIDVISGSVGDMAERGICEAFKVKQAVLLSS 1601
Query: 512 ADAVCTVLRVDQVI 525
+A +LRVD++I
Sbjct: 1602TEAAEMILRVDEII 1643
>TC216674 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/F26K24_12
{Arabidopsis thaliana;} , partial (42%)
Length = 1008
Score = 95.9 bits (237), Expect = 4e-20
Identities = 57/199 (28%), Positives = 99/199 (49%), Gaps = 3/199 (1%)
Frame = +2
Query: 330 TGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILDD 389
TG ++ + LG + +VG I G + +T+VLRG D +++
Sbjct: 14 TGGTVQTSVNNIIDEVLGTCEIFEERQVGNERFNIFNGCPSGQT-ATIVLRGGADQFIEE 190
Query: 390 LERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEM 449
ER++ D + + K+S V G A ++E+++ ++ + G Q I +A++ E+
Sbjct: 191 AERSLHDAIMIVRRALKNSTVVAGGGAIDMEISRYLRQHARTIAGKSQLFINSYAKALEV 370
Query: 450 IPRTLAENAGLNAMEIISSLYAEHA--SG-NTKVGIDLEEGVCKDVTTTHVWDLHVTKFF 506
IPR L +NAG +A ++++ L +HA SG G+D+ G D VW+ V K
Sbjct: 371 IPRQLCDNAGFDATDVLNKLRQKHALPSGEGAPYGVDIATGGIADSFANFVWEPAVVKIN 550
Query: 507 ALKYAADAVCTVLRVDQVI 525
A+ A +A C +L VD+ I
Sbjct: 551 AINAATEAACLILSVDETI 607
>TC227699 similar to UP|TCPZ_HUMAN (P40227) T-complex protein 1, zeta subunit
(TCP-1-zeta) (CCT-zeta) (CCT-zeta-1) (Tcp20) (HTR3),
partial (38%)
Length = 1136
Score = 94.4 bits (233), Expect = 1e-19
Identities = 74/274 (27%), Positives = 128/274 (46%), Gaps = 11/274 (4%)
Frame = +1
Query: 263 SAEQLENYSKTEEAKVEELIKAVADSGAKV---------IVSGGAVGEMALHFCERYKLM 313
SAEQ E E +V+E +K + + KV +++ + +L R ++
Sbjct: 7 SAEQREAMVAAERRQVDEKVKRIIELKNKVCSGNDSNFVVINQKGIDPPSLDLLAREGII 186
Query: 314 VLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNS 373
L+ + + + R G AV + P+ LG+ V +G T V+N + S
Sbjct: 187 ALRRAKRRNMERLVLACGGEAVNSVDDLTPECLGWAGLVYEHVLGEEKYTFVENVKNPFS 366
Query: 374 VSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKET 433
T++++G D + ++ AV DG+ K +D V GA A E+ + + + K+T
Sbjct: 367 C-TILIKGPNDHTIAQIKDAVRDGLRAVKNTLEDESVVLGAGAFEVAARQYLMN-EVKKT 540
Query: 434 --GLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKD 491
G Q + FA++ ++P+TLAEN+GL+ ++I +L EH GN VG+ L G D
Sbjct: 541 VQGRAQLGVEAFADALLVVPKTLAENSGLDTQDVIIALTGEHDKGNI-VGLSLNTGEPID 717
Query: 492 VTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
++D + K + V +L VD+VI
Sbjct: 718 PAMEGIFDNYSVKRQIINSGPVIVSQLLVVDEVI 819
>TC208068 homologue to PRF|2206327A.0|1587206|2206327A T complex protein.
{Cucumis sativus;} , partial (27%)
Length = 541
Score = 94.0 bits (232), Expect = 1e-19
Identities = 53/129 (41%), Positives = 77/129 (59%)
Frame = +1
Query: 15 KEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNE 74
+E L GLD A NI A K ++ I RTSLGP GM+KM+ + + +TND ATI+++
Sbjct: 154 QEQKSRLRGLD-AQKANISAGKAVARILRTSLGPKGMDKMLQSPDGDVTITNDGATILDQ 330
Query: 75 LEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKA 134
++V + AK++V ++Q EIGDG + AG LL+ AE L+ G+HP I GY A
Sbjct: 331 MDVDNQIAKLMVELSRSQDYEIGDGTTGVVVMAGALLEKAERLLERGIHPIRIAEGYEMA 510
Query: 135 IAKTIEILD 143
+E L+
Sbjct: 511 SRIAVEHLE 537
>TC228038 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {Arabidopsis
thaliana;} , partial (10%)
Length = 417
Score = 87.4 bits (215), Expect = 1e-17
Identities = 39/47 (82%), Positives = 43/47 (90%)
Frame = +3
Query: 479 KVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
KVGIDLEEG+CKDV+T +WDLHVTK FALKYAADA CTVLRVDQ+I
Sbjct: 3 KVGIDLEEGICKDVSTLSIWDLHVTKLFALKYAADAACTVLRVDQII 143
>TC213696 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/F26K24_12
{Arabidopsis thaliana;} , partial (21%)
Length = 440
Score = 83.6 bits (205), Expect = 2e-16
Identities = 42/109 (38%), Positives = 66/109 (60%)
Frame = +1
Query: 13 MLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIV 72
+LKEG G + ++ NI+AC ++ + RT+LGP GM+K++ + + ++ND ATI+
Sbjct: 115 LLKEGTDTSQGKPQ-LVSNINACTAVADVVRTTLGPRGMDKLIHDDKGTVTISNDGATIM 291
Query: 73 NELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMG 121
L++ HPAAKIL K+Q E+GDG + A E L+ A+ I G
Sbjct: 292 KLLDIVHPAAKILADIAKSQDSEVGDGTTTVVLLAAEFLREAKPFIEDG 438
>TC205278 similar to PRF|2206327A.0|1587206|2206327A T complex protein.
{Cucumis sativus;} , partial (41%)
Length = 915
Score = 80.9 bits (198), Expect = 1e-15
Identities = 52/198 (26%), Positives = 100/198 (50%), Gaps = 2/198 (1%)
Frame = +2
Query: 331 GAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVS-TVVLRGSTDSILDD 389
G V + + +P+ LG V + G ++ E NS + T+ +RG I+++
Sbjct: 59 GGXIVPRFQELSPEKLGKAGMVREKSFGTTKDRMLYIEHCANSRAVTIFIRGGNKMIIEE 238
Query: 390 LERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEM 449
+R++ D + + + +++ V G + EI + V+ + + G++QYAI F ++ E
Sbjct: 239 TKRSLHDALCVARNLIRNNSIVYGGGSAEISCSIAVEAAADRYPGVEQYAIRAFGDALEA 418
Query: 450 IPRTLAENAGLNAMEIISSLYAEH-ASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFFAL 508
IP LAEN+GL +E +S++ ++ N GID + D+ +V++ + K L
Sbjct: 419 IPMALAENSGLQPIETLSAVKSQQIKDNNPHFGIDCNDVGTNDMREQNVFETLIGKQQQL 598
Query: 509 KYAADAVCTVLRVDQVIN 526
A V +L++D VI+
Sbjct: 599 LLATQVVKMILKIDDVIS 652
>TC234884 similar to UP|TCPA_DROME (P12613) T-complex protein 1, alpha
subunit (TCP-1-alpha) (CCT-alpha), partial (27%)
Length = 673
Score = 77.4 bits (189), Expect = 1e-14
Identities = 44/123 (35%), Positives = 73/123 (58%), Gaps = 8/123 (6%)
Frame = +2
Query: 411 VPGAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLY 470
V G A E L+ +++F+ + +Q AIA+FA+S +IP+TL+ NA +A ++++ L
Sbjct: 17 VAGGGAVEAALSIYLENFATSLSSREQLAIAEFAKSLLVIPKTLSVNAAQDATDLVAKLR 196
Query: 471 AEHASGNTKV--------GIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVD 522
+ H S TKV G+DL EGV +D V + ++K +LK+A +A T+LR+D
Sbjct: 197 SYHNSSQTKVDHANLKWIGLDLTEGVVRDNRKAGVLEPAISKIKSLKFATEAAITILRID 376
Query: 523 QVI 525
+I
Sbjct: 377 DMI 385
>TC230613 homologue to UP|Q8L7N0 (Q8L7N0) TCP-1 chaperonin-like protein
(At5g16070), partial (22%)
Length = 446
Score = 74.3 bits (181), Expect = 1e-13
Identities = 35/98 (35%), Positives = 58/98 (58%)
Frame = +3
Query: 27 AVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILV 86
A+ NI+A K L + +T+LGP G KM++ + +T D T++ E+++Q+P A ++
Sbjct: 150 ALHMNINAAKGLQDVLKTNLGPKGTIKMLVGGAGDIKLTKDGNTLLKEMQIQNPTAIMIA 329
Query: 87 LAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHP 124
AQ + GDG T+ F GEL++ +E I G+HP
Sbjct: 330 RTAVAQDDASGDGTTSTVIFIGELMKQSERYIDEGMHP 443
>TC227698 similar to UP|TCPZ_HUMAN (P40227) T-complex protein 1, zeta subunit
(TCP-1-zeta) (CCT-zeta) (CCT-zeta-1) (Tcp20) (HTR3),
partial (17%)
Length = 713
Score = 72.0 bits (175), Expect = 6e-13
Identities = 51/165 (30%), Positives = 85/165 (50%), Gaps = 2/165 (1%)
Frame = +1
Query: 363 TIVKNEEGGNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELA 422
T V+N + S T++++G D + ++ AV DG+ K +D V GA A E+
Sbjct: 7 TFVENVKNPFSC-TILIKGPNDHTIAQIKDAVRDGLRAVKNTLEDESVVLGAGAFEVAAR 183
Query: 423 KRVKDFSFKET--GLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKV 480
+ + + K+T G Q + FA++ ++P+TLAEN+GL+ ++I +L EH GN V
Sbjct: 184 QYLMN-EVKKTVQGRAQLGVEAFADALLVVPKTLAENSGLDTQDVIIALTGEHDKGNI-V 357
Query: 481 GIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
G+ L G D ++D + K + V +L VD+VI
Sbjct: 358 GLSLNTGEPIDPAMEGIFDNYSVKRQIINSGPVIVSQLLVVDEVI 492
>BE803835 similar to GP|24371053|db t-complex polypeptide 1 {Bruguiera
sexangula}, partial (32%)
Length = 541
Score = 70.9 bits (172), Expect = 1e-12
Identities = 44/158 (27%), Positives = 84/158 (52%), Gaps = 8/158 (5%)
Frame = +2
Query: 376 TVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKDFSFKETGL 435
T++LRG+ D +LD+++RA+ D ++ K + + V A E L+ ++ +
Sbjct: 35 TLILRGANDHMLDEMDRALHDALSIVKRTLESNTVVSAGGAVES*LSVYLEYLATPVGST 214
Query: 436 DQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTK--------VGIDLEEG 487
+Q AIA+FAES +I L+ NA +A E+++ L A H S ++ +G+DL +G
Sbjct: 215 EQLAIAEFAESLLIISNVLSVNAANDATELVAKLQAYHHSVQSRADKKHLSTMGLDLSQG 394
Query: 488 VCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
++ V + ++ ++ A +A T+L +D +I
Sbjct: 395 KIQNNLDAGVIEPAMSTVKIIQCATEAAITILTIDDMI 508
>TC205279 homologue to PRF|2206327A.0|1587206|2206327A T complex protein.
{Cucumis sativus;} , partial (52%)
Length = 838
Score = 67.8 bits (164), Expect = 1e-11
Identities = 50/238 (21%), Positives = 105/238 (44%), Gaps = 3/238 (1%)
Frame = +3
Query: 179 TICSLVADACIQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDA--VGSIKRI 236
++ + A + V + N+D ++V +GG L ++ ++ G+V+ D K+I
Sbjct: 123 SLAEIAVKAVLAVADLARKDVNLDLIKVEGKVGGKLEDTELIYGIVVDKDMSHPQMPKQI 302
Query: 237 EKAKVAVFVSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSG 296
E AK+A+ + TK V I + E+ + E+ +++++ D GA +++
Sbjct: 303 EDAKIAILTCPFEPPKPKTKHKVDIDTVEKFQTLRLQEQKYFDDMVQKCKDVGATLVICQ 482
Query: 297 GAVGEMALHFCERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEE 356
+ A H L ++ EL TG V + + +P+ LG V +
Sbjct: 483 WGFDDEANHLLMHRNLPAVRWVGGVELELIAIATGGRIVPRFQELSPEKLGKAGMVREKS 662
Query: 357 VGGVTVTIVKNEEGGNS-VSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPG 413
G ++ E NS T+ +RG I+++ +R++ D + + + +++ V G
Sbjct: 663 FGTTKDRMLYIEHCANSRAVTIFIRGGNKMIIEETKRSLHDALCVARNLIRNNSIVYG 836
>TC226488 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1, partial
(20%)
Length = 672
Score = 60.8 bits (146), Expect = 1e-09
Identities = 34/98 (34%), Positives = 58/98 (58%), Gaps = 8/98 (8%)
Frame = +2
Query: 436 DQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTK--------VGIDLEEG 487
+Q AIA+FAES +IP+ L+ NA +A E+++ L A H S TK +G+DL +G
Sbjct: 8 EQLAIAQFAESLLIIPKVLSVNAAKDATELVAKLRAYHHSAQTKADKKHLSSMGLDLSQG 187
Query: 488 VCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
++ V + ++K +++A +A T+LR+D +I
Sbjct: 188 KIRNNLEAGVIEPAMSKVKIIQFATEAAITILRIDDMI 301
>TC215027 weakly similar to UP|Q8WSY6 (Q8WSY6) CCT chaperonin beta subunit,
partial (20%)
Length = 695
Score = 59.7 bits (143), Expect = 3e-09
Identities = 36/108 (33%), Positives = 55/108 (50%)
Frame = +3
Query: 418 EIELAKRVKDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAEHASGN 477
E+ +AK V + K G AI F+ + IP +A+NAGL++ E+IS L AEH
Sbjct: 15 EMVMAKEVDALAKKTPGKKSLAIEAFSRALLAIPTIIADNAGLDSAELISQLRAEHQKEG 194
Query: 478 TKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVI 525
GID+ G D+ + + K L + +A +LRVD++I
Sbjct: 195 CTSGIDVISGSVGDMAERGISEAFKVKQAVLLSSTEAAEMILRVDEII 338
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,682,969
Number of Sequences: 63676
Number of extensions: 173974
Number of successful extensions: 883
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 869
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 871
length of query: 528
length of database: 12,639,632
effective HSP length: 102
effective length of query: 426
effective length of database: 6,144,680
effective search space: 2617633680
effective search space used: 2617633680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0023.15


