

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0020.1
(384 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
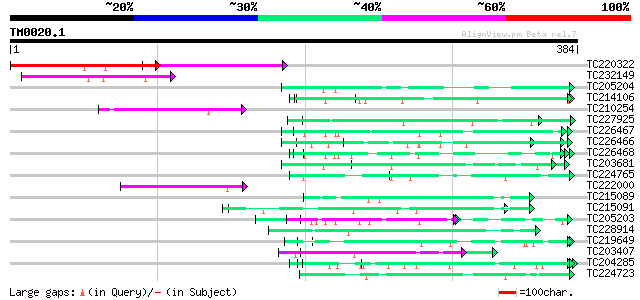
Score E
Sequences producing significant alignments: (bits) Value
TC220322 114 7e-26
TC232149 63 3e-10
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 62 3e-10
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 62 4e-10
TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F1... 60 1e-09
TC227925 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein ... 59 3e-09
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 59 5e-09
TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {... 57 1e-08
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 56 3e-08
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 56 3e-08
TC224765 similar to UP|Q09082 (Q09082) Extensin (Class I), parti... 56 3e-08
TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transp... 56 3e-08
TC215089 55 5e-08
TC215091 UP|CUT2_CAEEL (P34682) Cuticlin 2 precursor, partial (10%) 54 1e-07
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 53 3e-07
TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (... 53 3e-07
TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich... 52 3e-07
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 52 3e-07
TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 52 4e-07
TC224723 hydroxyproline-rich glycoprotein 52 6e-07
>TC220322
Length = 1264
Score = 114 bits (285), Expect = 7e-26
Identities = 62/106 (58%), Positives = 70/106 (65%), Gaps = 4/106 (3%)
Frame = +2
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKE--AAQKWTATW 58
IL+YR KPIITMLEEIR Y+MRT+A K+KL G G L VQ RLEKE A +WT W
Sbjct: 188 ILQYRCKPIITMLEEIRSYIMRTMAARKVKLSGKPGPLCLVQYKRLEKEFHFANQWTPIW 367
Query: 59 SGQES--RFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAI 102
G R+EV WG +V V+LA TCTC WQL GMPC HA A I
Sbjct: 368 CGDNMGLRYEVHMWGNKVEVNLAGWTCTCGVWQLTGMPCRHAIATI 505
Score = 77.0 bits (188), Expect = 1e-14
Identities = 37/98 (37%), Positives = 51/98 (51%)
Frame = +1
Query: 91 NGMPCFHACAAIGWKHERPENYVHAWLTMGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPP 150
+ MP H C K +PE+ H WL++ AY TY+ I+P + QYW T Y P+PP
Sbjct: 475 DAMPTCH-CNNNSHKGGKPEDMCHEWLSIEAYNKTYQHFIEPVQGPQYWAQTQYTHPVPP 651
Query: 151 NMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGR 188
+ + GRPKKNR+R E+ V G + GR
Sbjct: 652 YKRVQRGRPKKNRKRSVDEDNVTGHKLKRKLAEFTCGR 765
>TC232149
Length = 1005
Score = 62.8 bits (151), Expect = 3e-10
Identities = 45/138 (32%), Positives = 64/138 (45%), Gaps = 34/138 (24%)
Frame = -3
Query: 9 IITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQ--KWTATWSGQE--SR 64
I+ M E+IR Y+M + ++K+K+ L P+Q+SR+EK + K TA W G +R
Sbjct: 427 ILNMCEDIRTYIMCKMKSDKLKMATRLRPLVPMQQSRIEKGKVESNK*TANWVGDPDGTR 248
Query: 65 FEVTCWGQRVAVDLAAGTCTCRWWQL------------------------------NGMP 94
F+V + R+ VDL + TCR WQL GMP
Sbjct: 247 FKVCHYDTRLNVDLQN*SFTCRMWQLIGIPLS*LNLSILDIMYCVTMNLMFFWFIYIGMP 68
Query: 95 CFHACAAIGWKHERPENY 112
C HA AI + + PE Y
Sbjct: 67 CRHAIVAIRYNYHIPEGY 14
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 62.4 bits (150), Expect = 3e-10
Identities = 58/205 (28%), Positives = 67/205 (32%), Gaps = 7/205 (3%)
Frame = +1
Query: 185 LAGRGPTGSQTPLAPAAAAASYPTITT-----PQGPPPLQ--PRVAYRAPIAAAAAAPRQ 237
+AG G G T AP +S P I + P PPP Q P + P ++ P
Sbjct: 160 VAGVGAQGPSTSPAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPAS 339
Query: 238 PPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVV 297
PPP P A P + P PPP P P PPP +P A PP P
Sbjct: 340 PPPSSPPPASPPPAS--PPPASPPPASP----PPASPPPASPPPATPPPASPPPFSP--- 492
Query: 298 YSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPP 357
P AT PP PA T P S SP P
Sbjct: 493 -----------------PPATPPPATPP--------------PALTPTPLSSPPATSPAP 579
Query: 358 GPATHELPGGPIVFMPTPSIQPRPP 382
PA P P PS+ P
Sbjct: 580 APAKVVAPALSPSLAPGPSLSTISP 654
Score = 36.2 bits (82), Expect = 0.026
Identities = 33/127 (25%), Positives = 43/127 (32%)
Frame = +1
Query: 253 VVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQR 312
VVA P A P+ PP ++ + PP Q S P +
Sbjct: 157 VVAGVGAQGPSTSPAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPA 336
Query: 313 GAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFM 372
P ++ +PP PP PA+ PP S SPPP P P F
Sbjct: 337 SPPPSSPPPASPPPAS----PPPASPPPASP--PPASPPPASPPPATPP---PASPPPFS 489
Query: 373 PTPSIQP 379
P P+ P
Sbjct: 490 PPPATPP 510
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 62.0 bits (149), Expect = 4e-10
Identities = 51/188 (27%), Positives = 64/188 (33%)
Frame = +1
Query: 195 TPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVV 254
+P P+ Y P PPP P R+P + P PPP P + P+
Sbjct: 16 SPPPPSPPPPVYSPPPPPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPVQYY 195
Query: 255 AAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGA 314
+ PPP P PPP P+ P P V+S P V +
Sbjct: 196 YS-SPPPPQHSPPPPPPHSPPPPPPSPPPPVYPYLSPPPPPPVHSPPPPVYSP---PPPP 363
Query: 315 PHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPT 374
P PP PP +EP P SPPP P+ H P + P
Sbjct: 364 PSPPPCIEPPP-------PPPPCIEPPPPPPSPPPCEEHSPPP-PSPHPAP-----YHPP 504
Query: 375 PSIQPRPP 382
PS P PP
Sbjct: 505 PSPSPPPP 528
Score = 52.4 bits (124), Expect = 3e-07
Identities = 45/148 (30%), Positives = 52/148 (34%)
Frame = +1
Query: 235 PRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQP 294
P PPP P Y P P P P P R PPP +P +PP P
Sbjct: 13 PSPPPPSPPPPVYSPP------PPPPSPPPPSPTYCVRSPPPPSP------PPPSPPPPP 156
Query: 295 RVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRS 354
V+S P V + H+ PP PP P + PP S
Sbjct: 157 PPVFSPPPPVQYYYSSPPPPQHS-----PPP-------PPPHSPPPPPPSPPPPVYPYLS 300
Query: 355 PPPGPATHELPGGPIVFMPTPSIQPRPP 382
PPP P H P P V+ P P P PP
Sbjct: 301 PPPPPPVHSPP--PPVYSPPPP-PPSPP 375
Score = 51.6 bits (122), Expect = 6e-07
Identities = 56/202 (27%), Positives = 67/202 (32%), Gaps = 9/202 (4%)
Frame = +1
Query: 190 PTGSQTPLAPAAAAASYPTITTP--QGPPPLQPRVAYRAPIAAAAAAP----RQPPPLQP 243
P S P +P S P + P PPP P + P+ ++P PPP P
Sbjct: 64 PPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPVQYYYSSPPPPQHSPPPPPP 243
Query: 244 RVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPS 303
P P PPP+ P + P PPP+ +S PP P P
Sbjct: 244 HSPPPPP------PSPPPPVYPYLSPPP--PPPVHSPPPPVYSPPPPPPSPPPCIEPPPP 399
Query: 304 VATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHE 363
I P PP PP P HPP S SPPP P +
Sbjct: 400 PPPCIEPPPPPP------SPPPCEEHSPPPPSPHPAP---YHPPPSP---SPPPPPVQYN 543
Query: 364 LPGGPIVFMPTPSI---QPRPP 382
P P PTP P PP
Sbjct: 544 SPPPPSPPPPTPVYHYNSPPPP 609
Score = 47.8 bits (112), Expect = 8e-06
Identities = 47/201 (23%), Positives = 70/201 (34%), Gaps = 13/201 (6%)
Frame = +1
Query: 194 QTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPP--PLQPRVAYRAPI 251
++P P+ S P P PP + Y +P + P PP P P + P+
Sbjct: 106 RSPPPPSPPPPSPPPPPPPVFSPPPPVQYYYSSPPPPQHSPPPPPPHSPPPPPPSPPPPV 285
Query: 252 AVVAAPRQPPPLQ--PRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIM 309
+P PPP+ P +P PPP P + PP P + P +
Sbjct: 286 YPYLSPPPPPPVHSPPPPVYSPPPPPPSPPPC-----IEPPPPPPPCIEPPPPPPSPPPC 450
Query: 310 NQRGAP---------HATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPA 360
+ P H + PP ++ PP P T + S S PP
Sbjct: 451 EEHSPPPPSPHPAPYHPPPSPSPPPPPVQYNSPPPPSPPPPTPVYHYNSPPPPSFPPPTP 630
Query: 361 THELPGGPIVFMPTPSIQPRP 381
+E P P++ + S P P
Sbjct: 631 VYEGPLPPVIGVSYASPPPPP 693
>TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17A9_9
{Arabidopsis thaliana;} , partial (6%)
Length = 814
Score = 60.5 bits (145), Expect = 1e-09
Identities = 39/105 (37%), Positives = 50/105 (47%), Gaps = 5/105 (4%)
Frame = +1
Query: 61 QESRFEVTCWGQRV-AVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
Q S FEV G+ V VD+ C+C+ WQL G+PC HA A P +Y + T+
Sbjct: 25 QGSTFEVR--GESVDIVDIDNWDCSCKGWQLTGVPCCHAIAVFECVGRSPYDYCSRYFTV 198
Query: 120 GAYRSTYEFAIQPT----RSQQYWEPTPYEKPIPPNMKKKAGRPK 160
YR TY +I P + E T IPP K+ GRPK
Sbjct: 199 ENYRLTYAESIHPVPNVDKPPVQGESTALVMVIPPPTKRPPGRPK 333
>TC227925 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein precursor,
partial (88%)
Length = 1035
Score = 59.3 bits (142), Expect = 3e-09
Identities = 56/205 (27%), Positives = 76/205 (36%), Gaps = 10/205 (4%)
Frame = +2
Query: 189 GPTGSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYR 248
GP S TP P++ AA+ P+ +P P P P A A AAP PP A
Sbjct: 137 GPASSPTPATPSSPAAASPSAPSPTNPSP-SPTPTGPAASGPAPAAPTPAPPSPAPAATA 313
Query: 249 APIAVVAAPRQP-PPLQP--RAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVA 305
A + AP P PP P R+ A PP A A++ ++ P + R VA
Sbjct: 314 AAASSATAPGAPHPPPSPSSRSTTAATTTPPTASASSTVLTSP*PSPRTRAKACALSWVA 493
Query: 306 TAIMNQR-------GAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPG 358
+ R P T + + F R TT+ S PP
Sbjct: 494 ATTFSPRVRACCSTVCPRVTDPWWLARAGARLF--TRTNSVAGTTSTTLTRARDLSTPPS 667
Query: 359 PATHELPGGPIVFMPTPSIQPRPPQ 383
+T LP ++ P PS PP+
Sbjct: 668 SSTRALPPLHLLMTPPPSCTSVPPR 742
Score = 51.2 bits (121), Expect = 8e-07
Identities = 52/167 (31%), Positives = 60/167 (35%), Gaps = 4/167 (2%)
Frame = +2
Query: 199 PAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPR 258
P A SY ++ PP PRV P + A P P +P A A+P
Sbjct: 23 PWLAHCSYAHSSSSLSPPFPHPRVLLS*P*STTATTPCGPASSPTPATPSSPAA--ASPS 196
Query: 259 QPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHAT 318
P P P + P P PA AA T APP + S ATA GAPH
Sbjct: 197 APSPTNPSPSPTPTGPAASGPAPAA--PTPAPPSPAPAATAAAASSATA----PGAPH-- 352
Query: 319 AAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGR----RSPPPGPAT 361
PP P R A TT PP + SP P P T
Sbjct: 353 ----PPP-------SPSSRSTTAATTTPPTASASSTVLTSP*PSPRT 460
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 58.5 bits (140), Expect = 5e-09
Identities = 63/208 (30%), Positives = 83/208 (39%), Gaps = 23/208 (11%)
Frame = +3
Query: 193 SQTPLA---PAAAAASYPTITTP----QGPPPLQPR---VAYRAPIAAAAAAPRQPPPLQ 242
S TPLA P AS PT +TP Q P + P+ V AP A A++P+ PPP
Sbjct: 111 SGTPLAVTLPPTVVASPPTTSTPPTTSQPPAIVAPKPAPVTPPAPKVAPASSPKAPPPQT 290
Query: 243 PRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPP---------LAPAAAASFSTQAP--- 290
P+ ++ V P PPP+ P + P PP PA A + +T AP
Sbjct: 291 PQ-PQPPKVSPVVTPTSPPPIPPPPVSTPPPLPPPKIAPTPAKTPPAPAPAKATPAPAPA 467
Query: 291 PLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQ 350
P +P + P T AP AP R R R P +
Sbjct: 468 PTKPAPTPAPVPPPPTL------APTPVVEVPAPAPSSHKHRRHRHRHRRHQAPAPAPTI 629
Query: 351 GRRSPP-PGPATHELPGGPIVFMPTPSI 377
R+SPP P +T E P P+PS+
Sbjct: 630 VRKSPPAPPDSTAESDTTP---APSPSL 704
Score = 58.5 bits (140), Expect = 5e-09
Identities = 51/202 (25%), Positives = 74/202 (36%), Gaps = 5/202 (2%)
Frame = +3
Query: 185 LAGRGPTGSQTPL-APAAAAASYPTITTPQGPPPLQ----PRVAYRAPIAAAAAAPRQPP 239
L G+ P+ +Q P APA + + P T+P PL P V P + QPP
Sbjct: 21 LCGQLPSNAQAPATAPANSPPTTPAATSPSSGTPLAVTLPPTVVASPPTTSTPPTTSQPP 200
Query: 240 PLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYS 299
+ +A AP PP + A++P+ PPP P Q P + P V +
Sbjct: 201 AI---------VAPKPAPVTPPAPKVAPASSPKAPPPQTP------QPQPPKVSPVVTPT 335
Query: 300 LTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGP 359
P + PPV PP + T P + + +P P P
Sbjct: 336 SPPPIPP-----------------PPVSTPPPLPPPKIAPTPAKTPPAPAPAKATPAPAP 464
Query: 360 ATHELPGGPIVFMPTPSIQPRP 381
A + P P P++ P P
Sbjct: 465 APTKPAPTPAPVPPPPTLAPTP 530
Score = 39.7 bits (91), Expect = 0.002
Identities = 41/134 (30%), Positives = 54/134 (39%), Gaps = 2/134 (1%)
Frame = +3
Query: 251 IAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAA--SFSTQAPPLQPRVVYSLTPSVATAI 308
+A+V+ Q P A AP PP PAA + S + A L P VV S P+ +T
Sbjct: 6 LALVSLCGQLPSNAQAPATAPANSPPTTPAATSPSSGTPLAVTLPPTVVAS-PPTTSTPP 182
Query: 309 MNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGP 368
+ P A A + PV PP +V PA S + PP P P
Sbjct: 183 TTSQ--PPAIVAPKPAPVT-----PPAPKVAPA-------SSPKAPPPQTPQPQPPKVSP 320
Query: 369 IVFMPTPSIQPRPP 382
+V +P P PP
Sbjct: 321 VVTPTSPPPIPPPP 362
>TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {Arabidopsis
thaliana;} , partial (22%)
Length = 978
Score = 57.4 bits (137), Expect = 1e-08
Identities = 60/219 (27%), Positives = 81/219 (36%), Gaps = 47/219 (21%)
Frame = +1
Query: 185 LAGRGPTGSQTPLAPAAAA----ASYPTITTPQG-----PPPLQPRVAYRAPIAAAAA-A 234
LA P+ P AA+ ++ PTI+ P P P+ P AP ++ A
Sbjct: 100 LAATSPSSGTPPAVTQAASPPTTSNPPTISQPPAIVAPKPAPVTPPAPKVAPASSPKAPT 279
Query: 235 PRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPP---------LAPAAAASF 285
P+ PPP P+V+ VAAP PPPL P A P PP +PA A +
Sbjct: 280 PQTPPPQPPKVS------PVAAPTLPPPLPPPPVATPPPLPPPKIAPTPAKTSPAPAPAK 441
Query: 286 STQAP------------PLQPRVVYSLTPSVATAI----------------MNQRGAPHA 317
+T AP P+ P + TP V +Q AP
Sbjct: 442 ATPAPAPAPTKPAPTPAPIPPPPTLAPTPIVEVPAPAPSSHKHRRHRHRHKRHQAPAPAP 621
Query: 318 TAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPP 356
T +++PP PP E TT P S + P
Sbjct: 622 TVIHKSPPA------PPDSTAESDTTPAPSPSLNLNAAP 720
Score = 53.9 bits (128), Expect = 1e-07
Identities = 53/184 (28%), Positives = 67/184 (35%), Gaps = 2/184 (1%)
Frame = +1
Query: 195 TPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVV 254
T A + ++ + P +T PP P A A P P P+VA A
Sbjct: 97 TLAATSPSSGTPPAVTQAASPPTTSNPPTISQPPAIVAPKPAPVTPPAPKVA-PASSPKA 273
Query: 255 AAPRQPPPLQPRAA--AAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQR 312
P+ PPP P+ + AAP PPPL P A+ PPL P P +A
Sbjct: 274 PTPQTPPPQPPKVSPVAAPTLPPPLPPPPVAT----PPPLPP-------PKIAPTPAKTS 420
Query: 313 GAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFM 372
AP A AP PA T P +P P P L PIV +
Sbjct: 421 PAPAPAKATPAPA--------------PAPTKPAP------TPAPIPPPPTLAPTPIVEV 540
Query: 373 PTPS 376
P P+
Sbjct: 541 PAPA 552
Score = 47.4 bits (111), Expect = 1e-05
Identities = 44/157 (28%), Positives = 58/157 (36%), Gaps = 2/157 (1%)
Frame = +1
Query: 227 PIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPP--PLAPAAAAS 284
P AA +P P P V A + P P QP A AP+ P P AP A +
Sbjct: 91 PTTLAATSPSSGTP--PAVTQAASPPTTSNP--PTISQPPAIVAPKPAPVTPPAPKVAPA 258
Query: 285 FSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTT 344
S +AP Q TP ++ AP PPV PP + T
Sbjct: 259 SSPKAPTPQ-------TPPPQPPKVSPVAAPTLPPPLPPPPVATPPPLPPPKIAPTPAKT 417
Query: 345 HPPQSQGRRSPPPGPATHELPGGPIVFMPTPSIQPRP 381
P + + +P P PA + P P P++ P P
Sbjct: 418 SPAPAPAKATPAPAPAPTKPAPTPAPIPPPPTLAPTP 528
Score = 37.7 bits (86), Expect = 0.009
Identities = 38/150 (25%), Positives = 48/150 (31%), Gaps = 19/150 (12%)
Frame = +1
Query: 252 AVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPL----------QPRVVYSLT 301
A + P PP A + PP AA+ +T PP +P V
Sbjct: 61 APASVPANSPPTTLAATSPSSGTPPAVTQAASPPTTSNPPTISQPPAIVAPKPAPVTPPA 240
Query: 302 PSVATAIMNQRGAPHATAAY--RAPPVRGKFFRPPRQRVEPATTTHPP-------QSQGR 352
P VA A + P + PV PP P T PP + +
Sbjct: 241 PKVAPASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLP--PPPVATPPPLPPPKIAPTPAK 414
Query: 353 RSPPPGPATHELPGGPIVFMPTPSIQPRPP 382
SP P PA P P P+ P PP
Sbjct: 415 TSPAPAPAKATPAPAPAPTKPAPTPAPIPP 504
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 55.8 bits (133), Expect = 3e-08
Identities = 59/197 (29%), Positives = 75/197 (37%), Gaps = 10/197 (5%)
Frame = -2
Query: 193 SQTPLA---PAAAAASYPTITTP----QGPPPLQPR---VAYRAPIAAAAAAPRQPPPLQ 242
S TPLA P AS PT +TP Q P + P+ V AP A A++P+ PPP
Sbjct: 908 SSTPLAVTQPPTVVASPPTTSTPPTTSQPPANVAPKPAPVKPSAPKVAPASSPKAPPPQI 729
Query: 243 PRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTP 302
P P +P PPPL P A+P PPL P A + PP +P
Sbjct: 728 P-----IPQPPKPSPVSPPPLPPPPVASP---PPLPPPKIAPTPAKTPP---------SP 600
Query: 303 SVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATH 362
+ A A AP +PA T P PP PA
Sbjct: 599 APAKATPAPAPAP----------------------TKPAPTPSPV--------PPPPAAT 510
Query: 363 ELPGGPIVFMPTPSIQP 379
P P++ +P P+ P
Sbjct: 509 PAPA-PVIEVPAPAPAP 462
Score = 53.1 bits (126), Expect = 2e-07
Identities = 55/189 (29%), Positives = 69/189 (36%), Gaps = 6/189 (3%)
Frame = -2
Query: 200 AAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAP-- 257
A A A+ P P PP P +P ++ A QPP VVA+P
Sbjct: 989 AQAPATVPVKLPPTTLPPTTPTAT--SPSSSTPLAVTQPP------------TVVASPPT 852
Query: 258 --RQPPPLQPRAAAAPRQPP--PLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRG 313
P QP A AP+ P P AP A + S +APP Q + PS
Sbjct: 851 TSTPPTTSQPPANVAPKPAPVKPSAPKVAPASSPKAPPPQIPIPQPPKPS---------- 702
Query: 314 APHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMP 373
P + PPV PP + T P + + +P P PA P P
Sbjct: 701 -PVSPPPLPPPPVASPPPLPPPKIAPTPAKTPPSPAPAKATPAPAPA-------PTKPAP 546
Query: 374 TPSIQPRPP 382
TPS P PP
Sbjct: 545 TPSPVPPPP 519
Score = 48.9 bits (115), Expect = 4e-06
Identities = 52/195 (26%), Positives = 72/195 (36%), Gaps = 8/195 (4%)
Frame = -2
Query: 190 PTGSQTPLAPAAAAASYPTITTPQGPPPL--QPRVAYRAPIAA---AAAAPRQPPPLQPR 244
P + P P A + S T PP + P P + A AP+ P P++P
Sbjct: 956 PPTTLPPTTPTATSPSSSTPLAVTQPPTVVASPPTTSTPPTTSQPPANVAPK-PAPVKPS 780
Query: 245 VAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSV 304
AP + AP P+ +P PPPL P AS PPL P P +
Sbjct: 779 APKVAPASSPKAPPPQIPIPQPPKPSPVSPPPLPPPPVAS----PPPLPP-------PKI 633
Query: 305 ATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHEL 364
A P + A +A P P +PA T P +P P P E+
Sbjct: 632 AP---TPAKTPPSPAPAKATPA------PAPAPTKPAPTPSPVPPPPAATPAPAPVI-EV 483
Query: 365 PG---GPIVFMPTPS 376
P P++ +P P+
Sbjct: 482 PAPAPAPVIEVPAPA 438
Score = 38.1 bits (87), Expect = 0.007
Identities = 35/127 (27%), Positives = 47/127 (36%), Gaps = 1/127 (0%)
Frame = -2
Query: 190 PTGSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRA 249
PT ++TP +PA A A T P P +P AA P P ++ A
Sbjct: 629 PTPAKTPPSPAPAKA-----TPAPAPAPTKPAPTPSPVPPPPAATPAPAPVIEVPAPAPA 465
Query: 250 PIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQP-RVVYSLTPSVATAI 308
P+ V AP R R APA A + ++PP P S T +
Sbjct: 464 PVIEVPAPAPAHHRHRRHRHRHRHRRHQAPAPAPTIIRKSPPAPPDSTAESDTAPAPSPS 285
Query: 309 MNQRGAP 315
+N AP
Sbjct: 284 LNLNAAP 264
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 55.8 bits (133), Expect = 3e-08
Identities = 52/199 (26%), Positives = 76/199 (38%), Gaps = 13/199 (6%)
Frame = +3
Query: 185 LAGRGPTGSQTPLAPAAAAASYPTITT----PQGPPPLQPRVAYRAPIAAAAAAPRQPPP 240
+ G+ P S T PAA + + T P+ P P+ + P ++ AA PP
Sbjct: 66 VGGQSPAASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAATATPPA 245
Query: 241 LQPRVAYRAPIAVVAAP------RQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQP 294
P VA A AP PP P AA P P +P A S+ P+
Sbjct: 246 SSPTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAPVPV 425
Query: 295 RVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRS 354
+L P+ ++ AP A AP + K + ++ PA + P G +
Sbjct: 426 SSPPALAPTTPAPVV----APSAEVPAPAPKSKKK-TKKSKKHTAPAPS---PSLLGPPA 581
Query: 355 PP---PGPATHELPGGPIV 370
PP PG + + GP V
Sbjct: 582 PPVGAPGSSQDSMSPGPAV 638
Score = 46.6 bits (109), Expect = 2e-05
Identities = 39/148 (26%), Positives = 54/148 (36%)
Frame = +3
Query: 232 AAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPP 291
AA+P PP + +P + P+ P P+ +++P PA++ + +T PP
Sbjct: 84 AASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSP-------PASSPNAATATPP 242
Query: 292 LQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQG 351
V S PS A A AP AT PP PP T P +
Sbjct: 243 ASSPTVAS-PPSKAAA-----PAPVATPPAATPPAA----TPPAATPPAVTPVSSPPAPV 392
Query: 352 RRSPPPGPATHELPGGPIVFMPTPSIQP 379
S PP P P P P + P
Sbjct: 393 PVSSPPAPVPVSSPPALAPTTPAPVVAP 476
Score = 29.3 bits (64), Expect = 3.1
Identities = 23/82 (28%), Positives = 34/82 (41%)
Frame = +1
Query: 198 APAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAP 257
AP AS P+ ++P PP + + R P P P PL P+ P +++
Sbjct: 7 APGIGKASSPSQSSPLSSPPSEDSLQRRRPPLLLRLPP--PLPLHPQ-----PQPLLSQN 165
Query: 258 RQPPPLQPRAAAAPRQPPPLAP 279
+ L PR +PP L P
Sbjct: 166 HRRQLLLPRHHLHQLRPPMLQP 231
>TC224765 similar to UP|Q09082 (Q09082) Extensin (Class I), partial (46%)
Length = 708
Score = 55.8 bits (133), Expect = 3e-08
Identities = 52/204 (25%), Positives = 75/204 (36%), Gaps = 11/204 (5%)
Frame = +3
Query: 190 PTGSQTPL---APAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVA 246
P S TP +P + S P+ + PPP P Y++P PP P
Sbjct: 15 PPPSPTPYYYHSPPPPSPSPPSPYYYKSPPPPSPVYIYKSP-----------PPPSPTYV 161
Query: 247 YRAPIAVVAAP----RQPPPLQPRAAAA--PRQPPPLAPAAAASFSTQAPPLQPRVVYSL 300
Y++P V +P + PPP P+ + PPP +P + Q+PP
Sbjct: 162 YKSPPPPVKSPPYYYQSPPPPSPKPKPPYYYKSPPPPSPKPKPPYYYQSPP--------- 314
Query: 301 TPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPP--RQRVEPATTTHPPQSQGRRSPPPG 358
PS P Y++PP +PP P + + PP + PPP
Sbjct: 315 PPS---------PVPKPPYYYKSPPPPSPVPKPPYYYHSPPPPSPSPPPPYYYKSPPPPS 467
Query: 359 PATHELPGGPIVFMPTPSIQPRPP 382
P+ P P + P P PP
Sbjct: 468 PS----PPPPYYYKSPPPPSPSPP 527
Score = 42.0 bits (97), Expect = 5e-04
Identities = 32/125 (25%), Positives = 42/125 (33%)
Frame = +3
Query: 258 RQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHA 317
+ PPP P PPP + + PP P +Y P P
Sbjct: 6 KSPPPPSPTPYYYHSPPPPSPSPPSPYYYKSPPPPSPVYIYKSPP-----------PPSP 152
Query: 318 TAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSI 377
T Y++PP K PP P + P+ PP P+ P P PS
Sbjct: 153 TYVYKSPPPPVK--SPPYYYQSPPPPSPKPKPPYYYKSPPPPSPKPKPPYYYQSPPPPSP 326
Query: 378 QPRPP 382
P+PP
Sbjct: 327 VPKPP 341
Score = 30.8 bits (68), Expect = 1.1
Identities = 22/89 (24%), Positives = 33/89 (36%), Gaps = 10/89 (11%)
Frame = +3
Query: 213 QGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAP- 271
Q PPP P + P + P P P P + P +P PPP ++ P
Sbjct: 303 QSPPPPSP--VPKPPYYYKSPPPPSPVPKPPYYYHSPP---PPSPSPPPPYYYKSPPPPS 467
Query: 272 ---------RQPPPLAPAAAASFSTQAPP 291
+ PPP +P+ + +PP
Sbjct: 468 PSPPPPYYYKSPPPPSPSPPPPYYYHSPP 554
>TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transposase
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (12%)
Length = 617
Score = 55.8 bits (133), Expect = 3e-08
Identities = 29/93 (31%), Positives = 45/93 (48%), Gaps = 7/93 (7%)
Frame = +2
Query: 76 VDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTMGAYRSTYEFAIQPTRS 135
V++ + +C+CR WQLNG+PC HA AA+ + + T ++R TY I P
Sbjct: 20 VNIGSHSCSCRDWQLNGIPCSHAAAALISCRKDVYAFSQKCFTAASFRDTYAETIHPIPG 199
Query: 136 QQYWEPTPYEK-------PIPPNMKKKAGRPKK 161
+ W T PP +++ GRP+K
Sbjct: 200 KLEWSKTGNSSMDDNILVVRPPKLRRPPGRPEK 298
>TC215089
Length = 802
Score = 55.1 bits (131), Expect = 5e-08
Identities = 48/173 (27%), Positives = 66/173 (37%), Gaps = 17/173 (9%)
Frame = +1
Query: 200 AAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPL-----------QPRVAYR 248
A A+ P +T P GPP P RAPI+ + PP + PR
Sbjct: 112 ATASEPEPPVT-PSGPPKAGPP---RAPISTSGPPKEGPPRVLITTSRSPKAGPPRTLVT 279
Query: 249 A-----PIAVVAAPRQPPPLQPRAAAAP-RQPPPLAPAAAASFSTQAPPLQPRVVYSLTP 302
A P+ P+ PP P + + P ++ PP P + PP P P
Sbjct: 280 ASEPEPPVTTSGPPKAGPPRAPISTSRPPKEGPPRVPITTSGPPKAGPPRAPVTASEPKP 459
Query: 303 SVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSP 355
V + + G P A PP G PPR P TT+ PP++ R+P
Sbjct: 460 PVTPSGPPKAGQPRAPITTSGPPKAG----PPR---APITTSGPPKAGSPRAP 597
Score = 39.7 bits (91), Expect = 0.002
Identities = 44/170 (25%), Positives = 59/170 (33%), Gaps = 23/170 (13%)
Frame = +1
Query: 145 EKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAG----------------- 187
E P+ P+ KAG P+ G ++ R + S AG
Sbjct: 130 EPPVTPSGPPKAGPPRAPISTSGPPKEGPPRVLITTSRSPKAGPPRTLVTASEPEPPVTT 309
Query: 188 RGPTGSQTPLAPAAAAA----SYPTIT-TPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQ 242
GP + P AP + + P + T GPP P RAP+ A+ P P
Sbjct: 310 SGPPKAGPPRAPISTSRPPKEGPPRVPITTSGPPKAGPP---RAPVTASEPKPPVTPSGP 480
Query: 243 PRVAY-RAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPP 291
P+ RAPI PP P A PP A + A + PP
Sbjct: 481 PKAGQPRAPITTSG----PPKAGPPRAPITTSGPPKAGSPRAPITISGPP 618
Score = 32.0 bits (71), Expect = 0.48
Identities = 35/116 (30%), Positives = 41/116 (35%)
Frame = +1
Query: 143 PYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGRGPTGSQTPLAPAAA 202
P PI + K G P+ G + AG R T S+ P TP P A
Sbjct: 331 PPRAPISTSRPPKEGPPRVPITTSGPPK--AGPPRAPVTASE-----PKPPVTPSGPPKA 489
Query: 203 AASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPR 258
ITT GPP P RAPI + P RAPI + PR
Sbjct: 490 GQPRAPITT-SGPPKAGPP---RAPITTSGPPKAGSP--------RAPITISGPPR 621
>TC215091 UP|CUT2_CAEEL (P34682) Cuticlin 2 precursor, partial (10%)
Length = 913
Score = 53.9 bits (128), Expect = 1e-07
Identities = 56/227 (24%), Positives = 80/227 (34%), Gaps = 16/227 (7%)
Frame = +1
Query: 145 EKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGRGPTGSQTPLAPAAAAA 204
E P+ P+ KAG P+ + + GP + P AP
Sbjct: 166 EPPVTPSGPPKAGPPR----------------------ALITTSGPPKAGPPKAP----- 264
Query: 205 SYPTITTPQGPPPLQPRVAYRAPIAAA--------------AAAPRQPPPLQPRVAYRAP 250
+T + PP+ P RAPI+ + + P+ PP P A
Sbjct: 265 ----VTASEPEPPVTPSGPPRAPISTSGPLKAGPPRALITTSGPPKAGPPRAPVTASEPE 432
Query: 251 IAVVAAPRQPP--PLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAI 308
+ A+ +PP PL P P+ PP AP + PP P V +
Sbjct: 433 PPITASEPEPPVTPLGP-----PKAGPPRAPITTSGPPKAGPPRAPVTASEPESPVTPSG 597
Query: 309 MNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSP 355
+ G P A PP G PPR P TT+ PP++ R+P
Sbjct: 598 PPKAGPPRAPITTSGPPKAG----PPR---APITTSGPPKAGPPRAP 717
Score = 49.3 bits (116), Expect = 3e-06
Identities = 49/198 (24%), Positives = 63/198 (31%), Gaps = 8/198 (4%)
Frame = +1
Query: 149 PPNMKKKAGRPKKNRRRDGTEE---QVAGRARRDATESQLAGRGPTGSQTPLAPAAAAAS 205
PP A P+ G +G + + + GP + P AP A+
Sbjct: 250 PPKAPVTASEPEPPVTPSGPPRAPISTSGPLKAGPPRALITTSGPPKAGPPRAPVTASEP 429
Query: 206 YPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQP 265
P IT + PP+ P P+ PP RAPI P+ PP P
Sbjct: 430 EPPITASEPEPPVTP-----------LGPPKAGPP-------RAPITTSGPPKAGPPRAP 555
Query: 266 RAAAAPRQP-----PPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAA 320
A+ P P PP A A +T PP + G P A
Sbjct: 556 VTASEPESPVTPSGPPKAGPPRAPITTSGPP-------------------KAGPPRAPIT 678
Query: 321 YRAPPVRGKFFRPPRQRV 338
PP G PPR V
Sbjct: 679 TSGPPKAG----PPRAPV 720
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 52.8 bits (125), Expect = 3e-07
Identities = 40/124 (32%), Positives = 54/124 (43%), Gaps = 7/124 (5%)
Frame = +3
Query: 188 RGPTGSQTPLAPAAAA----ASYPTITTPQGPPPLQ--PRVAYRAPIAAAAAAPRQPPPL 241
+GP+ S P P A+ +S P + P PPP Q P + P ++ P PPP
Sbjct: 270 QGPSTSPAPPTPQASPPPVQSSPPPV--PSSPPPSQSPPPASTPPPSPLSSPPPSSPPPS 443
Query: 242 QPRVAYRAPIAVVAAPRQPPPLQPRAAAAP-RQPPPLAPAAAASFSTQAPPLQPRVVYSL 300
P + P + P PPP P ++ P PPP +P A PP P SL
Sbjct: 444 SPPPSSPPPAS--PPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPP--SL 611
Query: 301 TPSV 304
TP+V
Sbjct: 612 TPTV 623
Score = 47.8 bits (112), Expect = 8e-06
Identities = 52/188 (27%), Positives = 68/188 (35%), Gaps = 4/188 (2%)
Frame = +3
Query: 198 APAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQ--PRVAYRAPIAVVA 255
A A ++ P TPQ PP P + P+ ++ + PPP P +P
Sbjct: 261 AQAQGPSTSPAPPTPQASPP--PVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSP 434
Query: 256 APRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAP 315
P PPP P P PPP +P A+ + PP P +P AT P
Sbjct: 435 PPSSPPPSSP----PPASPPPSSPPPASPPPSSPPPSSPP---PFSPPPAT-------PP 572
Query: 316 HATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMP-- 373
AT P PP + PP S SP P P+ VF P
Sbjct: 573 PATPPPAVP--------PPSLTPTVTPLSSPPVS----SPAPSPSK--------VFAPAL 692
Query: 374 TPSIQPRP 381
+PS+ P P
Sbjct: 693 SPSLAPGP 716
Score = 46.2 bits (108), Expect = 2e-05
Identities = 42/154 (27%), Positives = 59/154 (38%), Gaps = 15/154 (9%)
Frame = +3
Query: 167 GTEEQVAGRARRDATESQLAGRGPTGSQTPLAPAAAAASY--PTITTP-----QGPPPLQ 219
G Q G + A + A P S P P++ S P +TP PPP
Sbjct: 252 GVGAQAQGPSTSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSS 431
Query: 220 PRVAYRAPIAAAAAAP--RQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPP- 276
P + P + A+P PPP P + P + P PPP P A P PP
Sbjct: 432 PPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSS--PPPFSPPPATPPPATPPPAVPPP 605
Query: 277 -----LAPAAAASFSTQAPPLQPRVVYSLTPSVA 305
+ P ++ S+ AP +L+PS+A
Sbjct: 606 SLTPTVTPLSSPPVSSPAPSPSKVFAPALSPSLA 707
Score = 39.7 bits (91), Expect = 0.002
Identities = 35/131 (26%), Positives = 46/131 (34%), Gaps = 2/131 (1%)
Frame = +3
Query: 251 IAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMN 310
+A V A Q P P PPP+ + S+ P P + PS ++
Sbjct: 246 VAGVGAQAQGPSTSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPP 425
Query: 311 QRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPP--GPATHELPGGP 368
P + PP PP PA+ PP S SPPP P P P
Sbjct: 426 SSPPPSSPPPSSPPPA-----SPPPSSPPPASP--PPSSPPPSSPPPFSPPPATPPPATP 584
Query: 369 IVFMPTPSIQP 379
+P PS+ P
Sbjct: 585 PPAVPPPSLTP 617
Score = 29.6 bits (65), Expect = 2.4
Identities = 27/101 (26%), Positives = 32/101 (30%)
Frame = +3
Query: 190 PTGSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRA 249
P S P +P ++ P P PPP P A P P PP+
Sbjct: 495 PPASPPPSSPPPSSPP-PFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPV-------- 647
Query: 250 PIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAP 290
P P AP P LAP S ST +P
Sbjct: 648 ---------SSPAPSPSKVFAPALSPSLAP--GPSLSTISP 737
>TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1),
partial (3%)
Length = 784
Score = 52.8 bits (125), Expect = 3e-07
Identities = 49/200 (24%), Positives = 72/200 (35%), Gaps = 16/200 (8%)
Frame = +3
Query: 176 ARRDATESQLAGRGPTGSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPI------- 228
A R T S L P S P +P + + + PT + P P P + R P
Sbjct: 99 ATRRTTTSTLPPASPRSSSPPSSPPSPSTT-PTSSAPGNAPHSSPSASKRRPTPSGPSSA 275
Query: 229 ---------AAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAP 279
++ AAP + P P + ++ A+P P P ++ P LA
Sbjct: 276 ASTSPKPTSTSSRAAPSKSPFTWPSASLGTSMSSPASPPPPAPNASTSSTTSAASPALAS 455
Query: 280 AAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVE 339
+AA + S P P + TP+ A + + +T+ P R RV
Sbjct: 456 SAANTASATTAPSLPSIPSRTTPTTARSTPSSSNPTSSTSPTAT---------PKRTRVS 608
Query: 340 PATTTHPPQSQGRRSPPPGP 359
T P S RS PP P
Sbjct: 609 SPT---PSSSSTSRSSPPSP 659
Score = 35.8 bits (81), Expect = 0.033
Identities = 47/202 (23%), Positives = 66/202 (32%), Gaps = 32/202 (15%)
Frame = +3
Query: 207 PTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVA-------YRAPIAVVAAPRQ 259
P P+ PPP + + R A PP PR + + +AP
Sbjct: 33 PRWRKPRAPPPHRSQTPTRTTTATRRTTTSTLPPASPRSSSPPSSPPSPSTTPTSSAPGN 212
Query: 260 PPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTP------SVATAIMNQRG 313
P P +A+ R+P P P++AAS S + R S +P S+ T++ +
Sbjct: 213 APHSSP--SASKRRPTPSGPSSAASTSPKPTSTSSRAAPSKSPFTWPSASLGTSMSSPAS 386
Query: 314 AP-------------------HATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRS 354
P ++AA A R P T P S S
Sbjct: 387 PPPPAPNASTSSTTSAASPALASSAANTASATTAPSLPSIPSRTTPTTARSTPSSSNPTS 566
Query: 355 PPPGPATHELPGGPIVFMPTPS 376
AT P V PTPS
Sbjct: 567 STSPTAT---PKRTRVSSPTPS 623
>TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich protein,
partial (37%)
Length = 750
Score = 52.4 bits (124), Expect = 3e-07
Identities = 56/190 (29%), Positives = 63/190 (32%), Gaps = 4/190 (2%)
Frame = +2
Query: 196 PLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVA 255
P P P P GPPPL P P A P PPP P +
Sbjct: 212 PSPPGPPPPGSPP*LPPPGPPPLGPAPLPGPPPPRPAPPPGPPPPGPTPPPGPPP*PPLP 391
Query: 256 APRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVV--YSLTPSVATAIMNQRG 313
P P P A P PPP PA S PP P + L S ++ G
Sbjct: 392 GPAPLPGSPPPGPAPPPGPPPPGPA-----SPPGPPP*PPLPGPAPLPGSPPPGPVSPPG 556
Query: 314 APHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQ--SQGRRSPPPGPATHELPGGPIVF 371
P PP+ G P PA+ PP +PPPGP P GP
Sbjct: 557 PPP*------PPLPGPALPPGPPPPGPASPPGPPP*LPPPDPAPPPGPPP*PPPTGPPPP 718
Query: 372 MPTPSIQPRP 381
PTP P P
Sbjct: 719 GPTPPPGPPP 748
Score = 48.1 bits (113), Expect = 6e-06
Identities = 59/202 (29%), Positives = 67/202 (32%), Gaps = 7/202 (3%)
Frame = +2
Query: 187 GRGPTGSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVA 246
G P+G P P P +P GPPP P + P PPPL P
Sbjct: 164 GPPPSGPAPPPGP-------PP*PSPPGPPP---------PGSPP*LPPPGPPPLGPAPL 295
Query: 247 YRAPIAVVAAPRQPPPLQPRAAAAPRQPPPL---APAAAASFSTQAPPLQPRVVYSLTPS 303
P A P PPP P P PPL AP + APP P +P
Sbjct: 296 PGPPPPRPAPPPGPPPPGPTPPPGPPP*PPLPGPAPLPGSPPPGPAPPPGPPPPGPASPP 475
Query: 304 VATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRR----SPPPGP 359
G P P+ G PP V P P G PPPGP
Sbjct: 476 ---------GPPP*PPLPGPAPLPGS---PPPGPVSPPGPPP*PPLPGPALPPGPPPPGP 619
Query: 360 ATHELPGGPIVFMPTPSIQPRP 381
A+ P GP +P P P P
Sbjct: 620 AS---PPGPPP*LPPPDPAPPP 676
Score = 43.1 bits (100), Expect = 2e-04
Identities = 50/180 (27%), Positives = 57/180 (30%), Gaps = 3/180 (1%)
Frame = +2
Query: 206 YPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQP 265
YP P PP L P + AP P P P P P P PPPL P
Sbjct: 125 YPP*GPPPKPPSLGPPPSGPAPPPGPPP*PSPPGPPPPGSPP*LP------PPGPPPLGP 286
Query: 266 RAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPP 325
P P P P PP P P + P A PP
Sbjct: 287 APLPGPPPPRPAPPPGPPPPGPTPPPGPPP-----*PPLPGPAPLPGSPPPGPAPPPGPP 451
Query: 326 VRGKFFRPPRQRVEPATTTHPP---QSQGRRSPPPGPATHELPGGPIVFMPTPSIQPRPP 382
PP P PP + SPPPGP + P P +P P++ P PP
Sbjct: 452 -------PPGPASPPGPPP*PPLPGPAPLPGSPPPGPVSPPGP-PP*PPLPGPALPPGPP 607
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 52.4 bits (124), Expect = 3e-07
Identities = 41/132 (31%), Positives = 57/132 (43%), Gaps = 5/132 (3%)
Frame = +2
Query: 183 SQLAGRGPTG--SQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAA--AAPRQP 238
+Q G P+ + TP P +A P TTP PPP P A P AA AA P
Sbjct: 188 AQAPGAAPSQPPTTTPSPPPPRSAPAPAPTTPATPPPATPPPAATPPPAATPPPAATPTP 367
Query: 239 PPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVY 298
P P A + A+P P P + +P PP P+ S S + PP P +
Sbjct: 368 APAPPTAAPTPASSPAASPPSPSPTVTPSPTSPNTPP--GPSPGPSGSAEPPP--PSAAF 535
Query: 299 SLTPS-VATAIM 309
S + + +AT+ +
Sbjct: 536 SASKAFIATSAL 571
Score = 46.6 bits (109), Expect = 2e-05
Identities = 45/133 (33%), Positives = 51/133 (37%)
Frame = +2
Query: 198 APAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAP 257
AP AA + PT TTP PPP PR A A A P PPP P P A P
Sbjct: 194 APGAAPSQPPT-TTPS-PPP--PRSA----PAPAPTTPATPPPATP------PPAATPPP 331
Query: 258 RQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHA 317
PP P A P PP A AS +PP V TPS + +P
Sbjct: 332 AATPP--PAATPTPAPAPPTAAPTPASSPAASPPSPSPTV---TPSPTSPNTPPGPSPGP 496
Query: 318 TAAYRAPPVRGKF 330
+ + PP F
Sbjct: 497 SGSAEPPPPSAAF 535
Score = 40.8 bits (94), Expect = 0.001
Identities = 43/153 (28%), Positives = 54/153 (35%)
Frame = +2
Query: 230 AAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQA 289
A AAP QPP P PPP PR+A AP P P A
Sbjct: 194 APGAAPSQPPTTTP---------------SPPP--PRSAPAPAPTTPATPPPATPPPAAT 322
Query: 290 PPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQS 349
PP + TP+ A A P ++ A P P V P+ T+
Sbjct: 323 PPPAATPPPAATPTPAPAPPTAAPTPASSPAASPP--------SPSPTVTPSPTS----- 463
Query: 350 QGRRSPPPGPATHELPGGPIVFMPTPSIQPRPP 382
+ PPGP+ PG P+ S +P PP
Sbjct: 464 ---PNTPPGPS----PG------PSGSAEPPPP 523
>TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (32%)
Length = 1539
Score = 52.0 bits (123), Expect = 4e-07
Identities = 48/191 (25%), Positives = 69/191 (35%), Gaps = 7/191 (3%)
Frame = +1
Query: 199 PAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPR 258
P P P+ PPP P+V P +PPP++P + P+ P+
Sbjct: 178 PPPVPEKKPPPPVPEKPPPCPPKVV----------PPPKPPPVKPPP--KPPVKPPCPPK 321
Query: 259 QPPPLQPR--AAAAPRQP-PPLAPAAAASFSTQAPPLQPRV--VYSLTPSVATAIMNQRG 313
PP P+ P P PP P + PP P+V + P +
Sbjct: 322 VEPPHPPKPPKVKPPHPPHPPKKPPSPPKVKPPHPPKPPKVEPPHPPKPPKKPPCPPKVK 501
Query: 314 APHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGP--ATHELPGGPIVF 371
PH + P +PP++ P P + ++PPP P E P P
Sbjct: 502 PPHPPKPPKVEPPHPP--KPPKKPPSPPKVVPPLPPKPPKNPPPLPPKPPKEPPTPPKEP 675
Query: 372 MPTPSIQPRPP 382
PTP +P PP
Sbjct: 676 PPTPPKKPCPP 708
Score = 50.4 bits (119), Expect = 1e-06
Identities = 48/204 (23%), Positives = 64/204 (30%), Gaps = 11/204 (5%)
Frame = +1
Query: 190 PTGSQTPLAPAAAAASYPTITTPQGPPPLQP--RVAYRAPIAAAAAAPRQP--------- 238
P P P P + P PPP++P + + P P P
Sbjct: 190 PEKKPPPPVPEKPPPCPPKVVPPPKPPPVKPPPKPPVKPPCPPKVEPPHPPKPPKVKPPH 369
Query: 239 PPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVY 298
PP P+ P P +PP ++P P + PP P + P ++P
Sbjct: 370 PPHPPKKPPSPPKVKPPHPPKPPKVEPPHPPKPPKKPPCPPKVKPPHPPKPPKVEP---- 537
Query: 299 SLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPG 358
P P P + PP+ EP T PP + PPP
Sbjct: 538 ---PHPPKPPKKPPSPPKVVPPLPPKPPKNPPPLPPKPPKEPPT---PP-----KEPPPT 684
Query: 359 PATHELPGGPIVFMPTPSIQPRPP 382
P P PI P P I PP
Sbjct: 685 PPKKPCP--PIFKKPLPPIPKLPP 750
Score = 47.4 bits (111), Expect = 1e-05
Identities = 50/197 (25%), Positives = 69/197 (34%), Gaps = 11/197 (5%)
Frame = +1
Query: 196 PLAPAAAAASYPTITTPQGP------PPLQPRVAYRAPIAAAAAAPR--QPPPLQPRVAY 247
P P S P + P P PP P+ + P P +PP ++P
Sbjct: 370 PPHPPKKPPSPPKVKPPHPPKPPKVEPPHPPKPPKKPPCPPKVKPPHPPKPPKVEPPHPP 549
Query: 248 RAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATA 307
+ P + P+ PPL P+ P+ PPPL P + PP P+ P
Sbjct: 550 KPPKKPPSPPKVVPPLPPK---PPKNPPPLPPK-----PPKEPPTPPKEPPPTPPKKPCP 705
Query: 308 IMNQRGAPHATAAYRAPPVR-GKFFRPPRQRVEPATTTHPPQSQGRRSP--PPGPATHEL 364
+ ++ P PPV K PP + HP + P PP P L
Sbjct: 706 PIFKKPLPPIPKLPPLPPVPIYKKPLPPLPPLPKLPPLHPVPIYKKPLPPLPPLPKFPPL 885
Query: 365 PGGPIVFMPTPSIQPRP 381
PI P P + P P
Sbjct: 886 HPVPIYKKPLPPLPPLP 936
Score = 43.1 bits (100), Expect = 2e-04
Identities = 40/178 (22%), Positives = 57/178 (31%), Gaps = 32/178 (17%)
Frame = +1
Query: 239 PPLQPRVAYRAPIAVVA--------------------APRQPPPLQPRAAAAPRQP---- 274
PPL P + ++ P V P +PPP P+ P+ P
Sbjct: 103 PPLPPLIFHKPPPPVYTPPPVPVYTPPPVPEKKPPPPVPEKPPPCPPKVVPPPKPPPVKP 282
Query: 275 ---PPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFF 331
PP+ P PP P+V P P + P + +
Sbjct: 283 PPKPPVKPPCPPKVEPPHPPKPPKV---KPPHPPHPPKKPPSPPKVKPPHPPKPPKVEPP 453
Query: 332 RPPRQRVEP-----ATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSIQPRPPQN 384
PP+ +P HPP+ P P + P P V P P P+PP+N
Sbjct: 454 HPPKPPKKPPCPPKVKPPHPPKPPKVEPPHPPKPPKKPPSPPKVVPPLP---PKPPKN 618
Score = 36.6 bits (83), Expect = 0.020
Identities = 38/137 (27%), Positives = 50/137 (35%), Gaps = 2/137 (1%)
Frame = +1
Query: 141 PTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGRGPTGSQTPLAPA 200
PTP +KP PP KK K + + L P PL P
Sbjct: 679 PTPPKKPCPPIFKKPLPPIPK----------LPPLPPVPIYKKPLPPLPPLPKLPPLHPV 828
Query: 201 AAAASYPTITTPQGP--PPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPR 258
P P P PPL P Y+ P+ P+ PP L P Y+ P+ +
Sbjct: 829 PIYKK-PLPPLPPLPKFPPLHPVPIYKKPLPPLPPLPKFPP-LHPVPIYKKPLPPLPPLP 1002
Query: 259 QPPPLQPRAAAAPRQPP 275
+ PPL+P P+ PP
Sbjct: 1003KLPPLKP-LPPLPKLPP 1050
>TC224723 hydroxyproline-rich glycoprotein
Length = 2345
Score = 51.6 bits (122), Expect = 6e-07
Identities = 54/193 (27%), Positives = 72/193 (36%), Gaps = 7/193 (3%)
Frame = +1
Query: 197 LAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPP---LQPRVAYRAPIAV 253
L P +A A Y + P P P Y +P PPP + P Y +P
Sbjct: 694 LVPNSARAPYYYKSPPPPSPSPPPPYYYHSP----------PPPKEHVHPPYYYHSP--- 834
Query: 254 VAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAP-PLQPRVVYSLTPSVATAIMNQR 312
P P P P +P P P P S P P P Y +P +
Sbjct: 835 -PPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPSPPPPYYYHSPPPPSP----- 996
Query: 313 GAPHATAAYRAPPVRGKFFRPP---RQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPI 369
+P + Y++PP PP + P+ T+HPP +SPPP P ++ P
Sbjct: 997 -SPPSPYYYKSPPPPSPSPPPPYYYQSPPPPSPTSHPPYYY--KSPPP-PTSYPPPPYHY 1164
Query: 370 VFMPTPSIQPRPP 382
V P PS P PP
Sbjct: 1165VSPPPPSPSPPPP 1203
Score = 37.7 bits (86), Expect = 0.009
Identities = 34/144 (23%), Positives = 55/144 (37%), Gaps = 1/144 (0%)
Frame = -3
Query: 160 KKNRRRDGTEEQVAGRARRDATESQLAGRGPTGSQTPLAPAAAAASYPTITTPQGPPPL- 218
++ R R+ E+ R R E + + P ++P P+ + +P P P
Sbjct: 657 ERERERERERERERERERERERERERSPPPPYYYKSPPPPSPSPPPPYYYQSPPPPSPTP 478
Query: 219 QPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLA 278
P Y++P PPP Y +P PPP + PPP +
Sbjct: 477 HPPYYYKSP----------PPPTSHPPPYHYVSPPPPSPSPPPPYHYTS------PPPPS 346
Query: 279 PAAAASFSTQAPPLQPRVVYSLTP 302
PA A + ++PP P +Y+ P
Sbjct: 345 PAPAPKYIYKSPP-PPVYIYASPP 277
Score = 36.2 bits (82), Expect = 0.026
Identities = 31/114 (27%), Positives = 44/114 (38%), Gaps = 4/114 (3%)
Frame = +1
Query: 193 SQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAP----RQPPPLQPRVAYR 248
S P +P+ + Y P P P P P + + P + PPP P
Sbjct: 976 SPPPPSPSPPSPYYYKSPPPPSPSPPPPYYYQSPPPPSPTSHPPYYYKSPPP--PTSYPP 1149
Query: 249 APIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTP 302
P V+ P PP P PPP +PA A + ++PP P +Y+ P
Sbjct: 1150 PPYHYVSPP--PPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPP-PPVYIYASPP 1302
Score = 34.3 bits (77), Expect = 0.097
Identities = 33/124 (26%), Positives = 43/124 (34%)
Frame = -3
Query: 258 RQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHA 317
R PPP + PPP +P+ + Q+PP P T PH
Sbjct: 582 RSPPP-----PYYYKSPPPPSPSPPPPYYYQSPP----------PPSPT--------PHP 472
Query: 318 TAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSI 377
Y++PP PP T+HPP PPP P+ P P + P
Sbjct: 471 PYYYKSPP-------PP--------TSHPPPYHYVSPPPPSPS----PPPPYHYTSPPPP 349
Query: 378 QPRP 381
P P
Sbjct: 348 SPAP 337
Score = 32.7 bits (73), Expect = 0.28
Identities = 24/65 (36%), Positives = 30/65 (45%), Gaps = 3/65 (4%)
Frame = -3
Query: 321 YRAPPVRGKFFRPP---RQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSI 377
Y++PP PP + P+ T HPP +SPPP P +H P V P PS
Sbjct: 558 YKSPPPPSPSPPPPYYYQSPPPPSPTPHPPYYY--KSPPP-PTSHP-PPYHYVSPPPPSP 391
Query: 378 QPRPP 382
P PP
Sbjct: 390 SPPPP 376
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,062,651
Number of Sequences: 63676
Number of extensions: 496624
Number of successful extensions: 15068
Number of sequences better than 10.0: 1357
Number of HSP's better than 10.0 without gapping: 6585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10282
length of query: 384
length of database: 12,639,632
effective HSP length: 99
effective length of query: 285
effective length of database: 6,335,708
effective search space: 1805676780
effective search space used: 1805676780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0020.1


