

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0018.6
(456 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
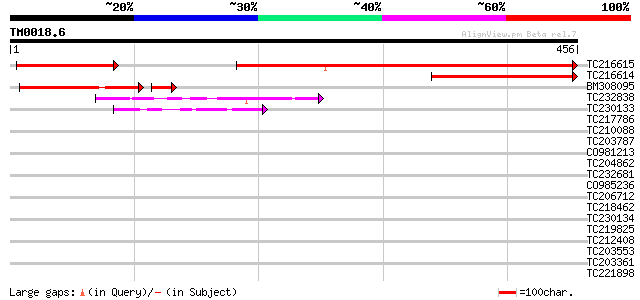
Score E
Sequences producing significant alignments: (bits) Value
TC216615 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At... 416 e-116
TC216614 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At... 210 1e-54
BM308095 similar to GP|22022558|gb| At5g12410 {Arabidopsis thali... 149 5e-39
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 44 2e-04
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 42 8e-04
TC217786 weakly similar to UP|Q6V9I6 (Q6V9I6) Histone deacetylas... 40 0.002
TC210088 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (4%) 40 0.003
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 40 0.003
CO981213 40 0.003
TC204862 similar to GB|AAM61695.1|21537354|AY085142 hydroxyproli... 39 0.004
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 39 0.005
CO985236 38 0.011
TC206712 similar to GB|AAT41855.1|48310625|BT014872 At2g19340 {A... 37 0.014
TC218462 37 0.018
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 37 0.024
TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process... 37 0.024
TC212408 similar to UP|Q867T7 (Q867T7) P67-like superoxide-gener... 36 0.041
TC203553 35 0.054
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 35 0.054
TC221898 similar to GB|AAS76268.1|45752740|BT012173 At4g23880 {A... 35 0.070
>TC216615 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At5g12410
{Arabidopsis thaliana;} , partial (62%)
Length = 1419
Score = 416 bits (1068), Expect = e-116
Identities = 209/277 (75%), Positives = 238/277 (85%), Gaps = 3/277 (1%)
Frame = +2
Query: 183 DEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDN 242
D+D E EGDE GDK PK+D+S ++N HD+ + K +PHK+D+ H ++T DN
Sbjct: 323 DDDDEEEEVQEGDEEKGDKKPKLDTSNDDNTSHDDGVPEKSDPHKVDEPHTQEVDETCDN 502
Query: 243 EGDNDNIKTK--TANELPVVKQCCKTNVPVQDSSNKVE-KSIDTLIDEELRELGDKKKRR 299
+G NDN+KT TA++LP VKQCCKTNVP + S+KVE KSID LI+EE +ELGDK KRR
Sbjct: 503 KGANDNLKTTKGTADDLPAVKQCCKTNVPACNFSDKVEHKSIDKLIEEEFKELGDKNKRR 682
Query: 300 FVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYAS 359
FVKL+SGCNGVVF+QMRKKDGD+SPKDIVHRIVTSAAST KHMSRFILR+LPIEVSCYAS
Sbjct: 683 FVKLDSGCNGVVFVQMRKKDGDRSPKDIVHRIVTSAASTGKHMSRFILRILPIEVSCYAS 862
Query: 360 KEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSVPGPHKVDLK 419
KEEISR IKPLVEQ FPV TQ PLKFAV+YEARANTG+DRMEIIDAVAKSVPGPHKVDLK
Sbjct: 863 KEEISRTIKPLVEQYFPVETQNPLKFAVLYEARANTGVDRMEIIDAVAKSVPGPHKVDLK 1042
Query: 420 NPDKTIVVEIARTVCLIGIIEKYKELSKYNLRELTSK 456
NPDKTIVVEIARTVCLIG+IEKYKELSKYNLR+LTS+
Sbjct: 1043NPDKTIVVEIARTVCLIGVIEKYKELSKYNLRQLTSR 1153
Score = 137 bits (344), Expect = 1e-32
Identities = 67/83 (80%), Positives = 74/83 (88%), Gaps = 1/83 (1%)
Frame = +3
Query: 6 KGDAAAAGGRKRKRYLPHNKPVKK-GSYPLHPGVQGVFITCEGGREHQASREALNILESF 64
K DAAAA G+KR+RYLPHNK VKK GSYPL PGVQG FITC+GGREHQASREALNIL+SF
Sbjct: 30 KPDAAAANGKKRRRYLPHNKAVKKKGSYPLRPGVQGFFITCDGGREHQASREALNILDSF 209
Query: 65 YEDLVDGEHLSVKELLNRPKNKK 87
YE+LVDGEH SVKEL ++P NKK
Sbjct: 210 YEELVDGEHSSVKELPSKPLNKK 278
>TC216614 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At5g12410
{Arabidopsis thaliana;} , partial (31%)
Length = 672
Score = 210 bits (534), Expect = 1e-54
Identities = 103/117 (88%), Positives = 112/117 (95%)
Frame = +3
Query: 340 KHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDR 399
+H SRFILR+LPIE++CYASKEEISRAIKPLVEQ FPV TQ P KFAV+YEARANTG+DR
Sbjct: 6 RHESRFILRILPIEIACYASKEEISRAIKPLVEQYFPVETQNPHKFAVLYEARANTGVDR 185
Query: 400 MEIIDAVAKSVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLRELTSK 456
MEIIDAVAKSVPGPHKVDLKNPDKTIVVEIARTVCLIG+IEKYKELSKYNLR+LTS+
Sbjct: 186 MEIIDAVAKSVPGPHKVDLKNPDKTIVVEIARTVCLIGVIEKYKELSKYNLRQLTSR 356
>BM308095 similar to GP|22022558|gb| At5g12410 {Arabidopsis thaliana},
partial (13%)
Length = 428
Score = 149 bits (377), Expect(2) = 5e-39
Identities = 76/100 (76%), Positives = 86/100 (86%), Gaps = 1/100 (1%)
Frame = +1
Query: 9 AAAAGGRKRKRYLPHNKPVKK-GSYPLHPGVQGVFITCEGGREHQASREALNILESFYED 67
A AA G+KR+RYLPHNK VKK GSYPL PGVQG FITC+GGREHQASREALNIL+SFYE+
Sbjct: 19 AVAANGKKRRRYLPHNKAVKKKGSYPLRPGVQGFFITCDGGREHQASREALNILDSFYEE 198
Query: 68 LVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEE 107
LVDGE KEL ++P NKKITFADSDSSSS +E++EEE
Sbjct: 199 LVDGE----KELPSKPLNKKITFADSDSSSSDGEEEEEEE 306
Score = 36.6 bits (83), Expect = 0.024
Identities = 14/24 (58%), Positives = 19/24 (78%)
Frame = +2
Query: 194 GDEANGDKMPKMDSSVNENAGHDN 217
GDE GDK PK+D+S ++NA HD+
Sbjct: 353 GDEEKGDKKPKLDTSSDDNASHDD 424
Score = 34.3 bits (77), Expect = 0.12
Identities = 14/20 (70%), Positives = 18/20 (90%)
Frame = +2
Query: 167 GDEAKGDEKPKLDSSVDEDA 186
GDE KGD+KPKLD+S D++A
Sbjct: 353 GDEEKGDKKPKLDTSSDDNA 412
Score = 30.0 bits (66), Expect(2) = 5e-39
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +2
Query: 115 GDEANGDKKLKLDSSVDEHA 134
GDE GDKK KLD+S D++A
Sbjct: 353 GDEEKGDKKPKLDTSSDDNA 412
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 43.9 bits (102), Expect = 2e-04
Identities = 47/189 (24%), Positives = 78/189 (40%), Gaps = 6/189 (3%)
Frame = +2
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSS 129
D E+ SV E+++ ++ + D D GDD DDEE+ G+ D D
Sbjct: 290 DEEYESVDEVVDDAEDDEDEEEDDDDDE-GDDNDDEEDDAPDGGDDD----------DDE 436
Query: 130 VDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIG 189
DE G ++ G E D+ D+++ D+ DE+ + + +ED G
Sbjct: 437 DDEEEGDVQR-----GGEPDDDDNDSDDDS-------DDDEDEDEEDEEEQGEEEDLGTE 580
Query: 190 ------EKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNE 243
E AE +EA+ D P+ + E D + E P K + + DD+
Sbjct: 581 YLIRPLETAEEEEASSDFEPEENGEEEEEDVDDEDCEKAEAPPK--RKRSDKDDSDDDDG 754
Query: 244 GDNDNIKTK 252
G++D +K
Sbjct: 755 GEDDERPSK 781
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 41.6 bits (96), Expect = 8e-04
Identities = 32/124 (25%), Positives = 57/124 (45%)
Frame = +3
Query: 84 KNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGA 143
K K + + D+S + DD+DD+ V +G++ N D++ E G
Sbjct: 240 KGKHTSEENKDASDTEDDDDDD-----NVNDGEDDNDDEE-------------DEDFSGD 365
Query: 144 NGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMP 203
+G E + DS D A GG ++ D+ DE D + ++D ++ EGD+ ++ P
Sbjct: 366 DGGE--EADSDDDPEANGGGGSDDDDGDDDED---DDNDEDDGDEDDEDEGDDDEDEESP 530
Query: 204 KMDS 207
+ S
Sbjct: 531 QPPS 542
Score = 32.3 bits (72), Expect = 0.46
Identities = 22/73 (30%), Positives = 33/73 (45%)
Frame = +3
Query: 175 KPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAA 234
K K S ++DA E + D+ D D +E+ D+ G E DD A
Sbjct: 240 KGKHTSEENKDASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDD---GGEEADSDDDPEAN 410
Query: 235 AGNKTDDNEGDND 247
G +DD++GD+D
Sbjct: 411 GGGGSDDDDGDDD 449
>TC217786 weakly similar to UP|Q6V9I6 (Q6V9I6) Histone deacetylase, partial
(48%)
Length = 1193
Score = 40.4 bits (93), Expect = 0.002
Identities = 35/150 (23%), Positives = 66/150 (43%)
Frame = +2
Query: 91 ADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPK 150
A S S S D++ D+ + + V ANG ++++ + + A ++ ANG ++
Sbjct: 335 AQSQSESDNDEDSDDFDEDIPVSA---ANGKPEIEVKNGIKPDANEAKQKKIANGEDEDS 505
Query: 151 LDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVN 210
+S D+++ T + + E D S DE+ EGD+ + ++ PK + N
Sbjct: 506 SESESDDDSSEDQLTANGDIESSEGD--DDSDDEE-------EGDDESDEETPKKIEASN 658
Query: 211 ENAGHDNEIGGKENPHKIDDVHAAAGNKTD 240
+ G D+ K+ P + KTD
Sbjct: 659 KR-GIDS---SKKTPVPVKKTKFVTPQKTD 736
Score = 33.1 bits (74), Expect = 0.27
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 6/63 (9%)
Frame = +2
Query: 76 VKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQ------VQVGEGDEANGDKKLKLDSS 129
+K N K KKI + + SS + +DD E Q ++ EGD+ + D++ D S
Sbjct: 443 IKPDANEAKQKKIANGEDEDSSESESDDDSSEDQLTANGDIESSEGDDDSDDEEEGDDES 622
Query: 130 VDE 132
+E
Sbjct: 623 DEE 631
>TC210088 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (4%)
Length = 683
Score = 39.7 bits (91), Expect = 0.003
Identities = 19/49 (38%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Frame = +1
Query: 92 DSDSSSSGDDEDDEEEVQV--QVGEGDEANGDKKLKLDSSVDEHAGIGE 138
D D + D E +EEE+ V + +G+E GD L D++ D++ GIGE
Sbjct: 82 DLDVEENIDAESEEEEIMVNFEASDGEEGEGDAPLPYDNNEDDYVGIGE 228
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 39.7 bits (91), Expect = 0.003
Identities = 30/123 (24%), Positives = 55/123 (44%), Gaps = 5/123 (4%)
Frame = -3
Query: 79 LLNRPKNK---KITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAG 135
L+N P K KI D + G+D +D+E G+G G+++L S ++ G
Sbjct: 697 LINTPTAKGESKIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEEL----SSEDGGG 530
Query: 136 IGEKAVGANGAEKPKLDSS--VDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAE 193
G + + ++K + +EN +GD+ D++ D +++AG E +
Sbjct: 529 YGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDED 350
Query: 194 GDE 196
G E
Sbjct: 349 GAE 341
>CO981213
Length = 710
Score = 39.7 bits (91), Expect = 0.003
Identities = 40/145 (27%), Positives = 59/145 (40%), Gaps = 18/145 (12%)
Frame = -1
Query: 115 GDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDEN--AGIGGKTEGDE--- 169
G E NG SS DE G+GE+ E +D VDE+ AG G +GDE
Sbjct: 533 GGELNG-------SSDDEGFGLGEEV*AEGVDEVRDVDFDVDEDVEAGDCGGVDGDEAAV 375
Query: 170 --------AKGDEKPKLDSS-----VDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHD 216
A+G +D++ V ED +G+ EG E G+ + ++ E H
Sbjct: 374 AVVDEEVGAEGGGSKVVDAASAVGDVGEDEAVGDAGEGGEDVGEWERVHEETLRELESHA 195
Query: 217 NEIGGKENPHKIDDVHAAAGNKTDD 241
G+ P + D+ G + D
Sbjct: 194 FCSRGEHAPDALVDLEVVVGREHRD 120
>TC204862 similar to GB|AAM61695.1|21537354|AY085142 hydroxyproline-rich
glycoprotein-like protein {Arabidopsis thaliana;} ,
partial (15%)
Length = 834
Score = 39.3 bits (90), Expect = 0.004
Identities = 38/134 (28%), Positives = 58/134 (42%), Gaps = 13/134 (9%)
Frame = -3
Query: 99 GDDEDDEEEVQVQVGEG----DEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSS 154
G DE +E E EG DEA G L + +VD+ G+GE+ +GA + +
Sbjct: 523 GADEGEEPECVHAEAEGLEFEDEAGGVVDLTVGGAVDDVVGVGEEGGEGSGAARVERAEG 344
Query: 155 VDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKA---------EGDEANGDKMPKM 205
E GG +EA+G + SSV E A + + EG E G+ +P+
Sbjct: 343 DLERGVEGGFAAIEEAEG-----VGSSVVEGANLAFEGFGFVVEVLLEGGEVVGELLPEE 179
Query: 206 DSSVNENAGHDNEI 219
+ AGH+ +
Sbjct: 178 GA-----AGHEGSL 152
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 38.9 bits (89), Expect = 0.005
Identities = 39/135 (28%), Positives = 59/135 (42%), Gaps = 1/135 (0%)
Frame = +1
Query: 94 DSSSSGDDEDDEEEVQVQ-VGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLD 152
D DDED+EEE VQ GE D+ + D + + DE E +G +P +
Sbjct: 265 DGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGEEADLGTEYLIRPLVT 444
Query: 153 SSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNEN 212
+ +E E +E +E+ +D DED GEK+E K D S +++
Sbjct: 445 AEEEE---ASSDFEPEENGEEEEEDVD---DED---GEKSEAPPKR--KRSDKDDSDDDD 591
Query: 213 AGHDNEIGGKENPHK 227
G D++ E P K
Sbjct: 592 GGEDDD----ERPSK 624
>CO985236
Length = 466
Score = 37.7 bits (86), Expect = 0.011
Identities = 44/158 (27%), Positives = 69/158 (42%), Gaps = 9/158 (5%)
Frame = -1
Query: 102 EDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGI 161
ED +E+ +V E + +G ++K D VD G + ++ + D VD +
Sbjct: 415 EDKKEDGVEEVNEDRKEDGVGEVKEDKEVD----------GVSCVKEVEEDGEVDGVKKV 266
Query: 162 GGKTEGDEAKGDEKPKLDSSV------DEDAGIGEKAEGDEANG---DKMPKMDSSVNEN 212
K +GDE K E+ K D + DED I E EG+ NG + P +D+ E
Sbjct: 265 EDK-KGDELKEIEEDKKDDHISENDKMDEDTEIKETMEGEPENGKEESEKPVVDAMEVEG 89
Query: 213 AGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIK 250
+ E KEN K++ V +++E D D K
Sbjct: 88 GIKEKE-ESKEN-EKVEVV-------MEEDEKDEDEDK 2
>TC206712 similar to GB|AAT41855.1|48310625|BT014872 At2g19340 {Arabidopsis
thaliana;} , partial (74%)
Length = 741
Score = 37.4 bits (85), Expect = 0.014
Identities = 44/141 (31%), Positives = 61/141 (43%), Gaps = 2/141 (1%)
Frame = -1
Query: 85 NKKITFADSDSSSSGD-DEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGA 143
N I++ S SGD D D+E V +G A G LD G +AV
Sbjct: 627 NSGISWIHYSSKISGDLDADEEHAHDVS*HDGGNAGGGVG-NLD---------GFRAVAI 478
Query: 144 NG-AEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKM 202
G E+ D++ DE+ GG+ D D D D AG+G + G +A G
Sbjct: 477 EGEVEENDADATEDEHES-GGEALDDVLAVDAAGHEDDGAD-GAGVGVLS-GADAGG--- 316
Query: 203 PKMDSSVNENAGHDNEIGGKE 223
+D V NAG D+E+G +E
Sbjct: 315 --LDDDVVNNAGDDHEVGEEE 259
>TC218462
Length = 1117
Score = 37.0 bits (84), Expect = 0.018
Identities = 38/152 (25%), Positives = 67/152 (44%), Gaps = 3/152 (1%)
Frame = +1
Query: 147 EKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLD-SSVDEDAGIGEKAEGDEANGDKMPKM 205
E P + VD+ A K +K DE P+ + +S + D + E DE ++M +
Sbjct: 220 ENPSEQNGVDQQA----KDNSVVSKTDENPRSEITSGENDQKVTENVGVDEEK-NEMKEA 384
Query: 206 DSSVNENAGHDNEIGGKENPHKID--DVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQC 263
+VNE DNE+ E D D+ A G +++DN G + TK +
Sbjct: 385 KETVNE---EDNEMKEAEETESEDNTDMKEAKGTESEDNNGMKEGKVTKDEEK------- 534
Query: 264 CKTNVPVQDSSNKVEKSIDTLIDEELRELGDK 295
K + S+ + ++D + E+L+ + +K
Sbjct: 535 -KQQAGEKFSAAAYKDNMDVVSREDLKSVFEK 627
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 36.6 bits (83), Expect = 0.024
Identities = 30/110 (27%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Frame = +2
Query: 80 LNRP-KNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVD---EHAG 135
L++P K K + + D+S + DD+DD++ V +G++ + D + D S D E A
Sbjct: 221 LDQPCKGKHTSEENKDASDTEDDDDDDD-----VNDGEDGDDDDEDDEDFSGDDGGEEAD 385
Query: 136 IGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED 185
+ G E D D++ + +GDE DE+ D DE+
Sbjct: 386 SDDDPEANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEE---DEDEDEE 526
Score = 34.3 bits (77), Expect = 0.12
Identities = 28/112 (25%), Positives = 43/112 (38%)
Frame = +2
Query: 137 GEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDE 196
G AN P +D + GK +E K D D D+D GE + D+
Sbjct: 161 GSLLTDANNLVSPMVDGRFPLDQPCKGKHTSEENK-DASDTEDDDDDDDVNDGEDGDDDD 337
Query: 197 ANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDN 248
+ + +G D G E DD A G ++DD++ D+D+
Sbjct: 338 EDDEDF----------SGDD----GGEEADSDDDPEANGGGESDDDDEDDDD 451
Score = 30.8 bits (68), Expect = 1.3
Identities = 24/118 (20%), Positives = 49/118 (41%)
Frame = +2
Query: 130 VDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIG 189
VD + + G + +E+ K S +++ +G++ D++ D S D+
Sbjct: 203 VDGRFPLDQPCKGKHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEA 382
Query: 190 EKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDND 247
+ + EANG G +++ +++ DD G++ DD+E D D
Sbjct: 383 DSDDDPEANG--------------GGESDDDDEDDDDDDDDNDEDDGDEDDDDEEDED 514
>TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process protein
MID2 (Serine-rich protein SMS1) (Protein kinase A
interference protein), partial (19%)
Length = 862
Score = 36.6 bits (83), Expect = 0.024
Identities = 34/104 (32%), Positives = 50/104 (47%), Gaps = 1/104 (0%)
Frame = -2
Query: 94 DSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDS 153
D S DDED EEE + VG E + D+ + + V++H E+ G E+
Sbjct: 330 D*KESEDDEDSEEEEEGVVGVDGEDHEDQGEEDEGVVEDH----EEGDP*EGEEEAL--- 172
Query: 154 SVDENAGIGGKTEGDEAKGDEKPKLDSSVDED-AGIGEKAEGDE 196
E+AG GG+ G+ GD ++ ED G GE+ EG+E
Sbjct: 171 ---EDAGDGGEGGGE---GDGLGGVEGGDGEDGGGGGEEGEGEE 58
>TC212408 similar to UP|Q867T7 (Q867T7) P67-like superoxide-generating NADPH
oxidase, partial (5%)
Length = 604
Score = 35.8 bits (81), Expect = 0.041
Identities = 30/112 (26%), Positives = 50/112 (43%), Gaps = 5/112 (4%)
Frame = -3
Query: 95 SSSSGDDEDDE--EEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLD 152
+S+ GDDED E+ + G +A +KL+ VD G E+ +E+ + D
Sbjct: 602 NSARGDDEDGGGVEDTEGYEDLGSDAQ-KRKLQGTDEVDYEDGPEEETHDGELSEEIEGD 426
Query: 153 ---SSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDK 201
S VD N T+ + ++G EKP ++DE + + + E K
Sbjct: 425 EDGSDVDANENYNNVTDANNSEGLEKPSKSKTIDEKQNLKREKKKSEPTTKK 270
>TC203553
Length = 660
Score = 35.4 bits (80), Expect = 0.054
Identities = 30/127 (23%), Positives = 53/127 (41%), Gaps = 2/127 (1%)
Frame = +3
Query: 61 LESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANG 120
++S + L D + L E + ++ S G + E ++ + +
Sbjct: 198 MQSTLDSLKDEQKLMESEF--EEQQNELRLMQEKGKSVGQGRSEIEALRDNLKHKEAEME 371
Query: 121 DKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEG--DEAKGDEKPKL 178
D K +L+ V++H I + V ANG + ++ DE +G K EG D +G K +L
Sbjct: 372 DLKRRLEIPVNDHPTIFPEIVTANGTIAAQDENGKDEISGESAKHEGDTDNNEGTIKSEL 551
Query: 179 DSSVDED 185
ED
Sbjct: 552 TKFAPED 572
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 35.4 bits (80), Expect = 0.054
Identities = 32/137 (23%), Positives = 55/137 (39%), Gaps = 14/137 (10%)
Frame = -1
Query: 79 LLNRPKNK---KITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAG 135
L N P K K D + G+D +D+E G+G G+++L S ++ G
Sbjct: 664 LTNTPTTKGERKTQSQDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEEL----SSEDGGG 497
Query: 136 IGEKAV-----------GANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDE 184
G + GA GAE+ + ++ + D+ DE+ + DE
Sbjct: 496 YGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDE 317
Query: 185 DAGIGEKAEGDEANGDK 201
D E+ E DE + ++
Sbjct: 316 DGAEDEENEEDEEDEEE 266
>TC221898 similar to GB|AAS76268.1|45752740|BT012173 At4g23880 {Arabidopsis
thaliana;} , partial (12%)
Length = 436
Score = 35.0 bits (79), Expect = 0.070
Identities = 26/88 (29%), Positives = 47/88 (52%), Gaps = 3/88 (3%)
Frame = +1
Query: 55 REALNILESFYEDLVDGEHLSVKE---LLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQ 111
+E L + ++ Y D E ++ K+ L ++ K + FADSD S G+D ++++ + +
Sbjct: 97 QEQLKVKKAVYRD----ELITKKKPPTLPSKAVKKSVRFADSDPSIFGEDHNEKKLAKGR 264
Query: 112 VGEGDEANGDKKLKLDSSVDEHAGIGEK 139
GDE G ++KL + DE A + K
Sbjct: 265 CSGGDELGG-IRVKLKMTKDEAARLLSK 345
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,087,051
Number of Sequences: 63676
Number of extensions: 198045
Number of successful extensions: 1478
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 1215
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1348
length of query: 456
length of database: 12,639,632
effective HSP length: 100
effective length of query: 356
effective length of database: 6,272,032
effective search space: 2232843392
effective search space used: 2232843392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0018.6


