

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
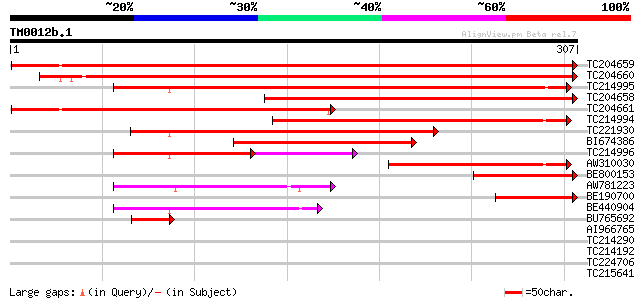
Query= TM0012b.1
(307 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204659 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isome... 533 e-152
TC204660 homologue to UP|TPIC_ARATH (Q9SKP6) Triosephosphate iso... 497 e-141
TC214995 UP|Q6GW08 (Q6GW08) Triosephosphate isomerase , complete 317 3e-87
TC204658 homologue to UP|TPIC_ARATH (Q9SKP6) Triosephosphate iso... 316 1e-86
TC204661 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isome... 280 4e-76
TC214994 homologue to UP|Q6GW08 (Q6GW08) Triosephosphate isomera... 211 4e-55
TC221930 similar to UP|Q6GW08 (Q6GW08) Triosephosphate isomerase... 144 4e-35
BI674386 121 3e-28
TC214996 UP|Q6GW08 (Q6GW08) Triosephosphate isomerase , partial... 97 9e-27
AW310030 similar to SP|P48491|TPIS_ Triosephosphate isomerase c... 115 2e-26
BE800153 homologue to SP|Q9SKP6|TPIC_ Triosephosphate isomerase ... 98 4e-21
AW781223 weakly similar to SP|P21820|TPIS Triosephosphate isomer... 92 3e-19
BE190700 similar to SP|Q9SKP6|TPIC_ Triosephosphate isomerase c... 68 6e-12
BE440904 similar to SP|P21820|TPIS_ Triosephosphate isomerase c... 67 1e-11
BU765692 similar to SP|Q9M4S8|TPIC_ Triosephosphate isomerase c... 47 1e-05
AI966765 weakly similar to SP|P21820|TPIS_ Triosephosphate isome... 37 0.009
TC214290 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp... 33 0.21
TC214192 similar to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp90... 32 0.28
TC224706 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp... 30 1.4
TC215641 homologue to UP|O04091 (O04091) Endomembrane protein EM... 29 3.1
>TC204659 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (90%)
Length = 1249
Score = 533 bits (1372), Expect = e-152
Identities = 264/306 (86%), Positives = 287/306 (93%)
Frame = +2
Query: 2 ATSLASQLSVGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAMAGSGKFFVG 61
+TSLASQL +GLRRP KLDS NSQS S+F NLRL +S +PSR+++AMAG+GKFFVG
Sbjct: 56 STSLASQLYIGLRRPCLKLDSFNSQS-FSVFDPNLRLSLSPPRPSRAIVAMAGTGKFFVG 232
Query: 62 GNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVS 121
GNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPF+YIDQVKNS+T+RIE+SAQNSWV
Sbjct: 233 GNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFLYIDQVKNSLTERIEISAQNSWVG 412
Query: 122 KGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGE 181
KGGAFTGEIS EQLKD G KWV+LGHSERRHIIGE DEFIGKKAAYAL++GLGVIACIGE
Sbjct: 413 KGGAFTGEISAEQLKDLGCKWVVLGHSERRHIIGENDEFIGKKAAYALSQGLGVIACIGE 592
Query: 182 LLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIR 241
LLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVA+P+QAQEVH A+R
Sbjct: 593 LLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVATPQQAQEVHVAVR 772
Query: 242 DWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVT 301
DWL+KNV EVASKTRIIYGGSVNGGNS+ELAK+EDIDGFLVGGASLKGPEFATI+NSVT
Sbjct: 773 DWLKKNVPDEVASKTRIIYGGSVNGGNSAELAKQEDIDGFLVGGASLKGPEFATIVNSVT 952
Query: 302 SKKVAA 307
SKKVAA
Sbjct: 953 SKKVAA 970
>TC204660 homologue to UP|TPIC_ARATH (Q9SKP6) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (83%)
Length = 1334
Score = 497 bits (1280), Expect = e-141
Identities = 255/297 (85%), Positives = 269/297 (89%), Gaps = 6/297 (2%)
Frame = +1
Query: 17 SPKLDSLNSQ---STHSLF---HSNLRLPISSSKPSRSVIAMAGSGKFFVGGNWKCNGTK 70
S L SLNSQ S+ S F HS L P SSKPSR V+AMAGSGKFFVGGNWKCNGTK
Sbjct: 121 SSNLHSLNSQPFSSSLSFFRNVHSTLSFP--SSKPSRGVVAMAGSGKFFVGGNWKCNGTK 294
Query: 71 DSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVSKGGAFTGEI 130
DSI KLV+DLNSA LE DVDVVVAPPFVYIDQVKNSITDRIE+SAQNSWV KGGAFTGEI
Sbjct: 295 DSIRKLVSDLNSATLESDVDVVVAPPFVYIDQVKNSITDRIEISAQNSWVGKGGAFTGEI 474
Query: 131 SVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELLEEREAGK 190
SVEQLKD G KWVILGHSERRH+IGE DEFIGKKA YAL+EGLGVIACIGELL+EREAGK
Sbjct: 475 SVEQLKDLGCKWVILGHSERRHVIGENDEFIGKKAVYALSEGLGVIACIGELLQEREAGK 654
Query: 191 TFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSA 250
TFDVCFQQLKA+AD VPSWDNIVIAYEPVWAIGTGKVA+PEQAQEVHAA+RDWL+KNVSA
Sbjct: 655 TFDVCFQQLKAFADVVPSWDNIVIAYEPVWAIGTGKVATPEQAQEVHAAVRDWLKKNVSA 834
Query: 251 EVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
EV+SKTRIIYGGSVN NS+ELAK+EDIDGFLVGGASLKGPEFATIINSVTSKKVAA
Sbjct: 835 EVSSKTRIIYGGSVNASNSAELAKQEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 1005
>TC214995 UP|Q6GW08 (Q6GW08) Triosephosphate isomerase , complete
Length = 1090
Score = 317 bits (813), Expect = 3e-87
Identities = 157/250 (62%), Positives = 190/250 (75%), Gaps = 2/250 (0%)
Frame = +2
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN AK+ E V+VVV+PPFV++ VK+ + VS
Sbjct: 101 KFFVGGNWKCNGTTEEVKKIVTTLNEAKVPGEDVVEVVVSPPFVFLPVVKSLLRPDFHVS 280
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGA+TGE+S E L + G+ WVI+GHSERR ++ E +EF+G K AYAL +GL
Sbjct: 281 AQNCWVRKGGAYTGEVSAEMLVNLGIPWVIIGHSERRQLLNESNEFVGDKVAYALQQGLK 460
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIACIGE LE+REAG T V +Q KA A + +WDN+V+AYEPVWAIGTGKVA+P QAQ
Sbjct: 461 VIACIGETLEQREAGTTTAVVAEQTKAIAAKISNWDNVVLAYEPVWAIGTGKVATPAQAQ 640
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +R W+ NVSAEVA+ RIIYGGSVNGGN ELA + D+DGFLVGGASLK EF
Sbjct: 641 EVHADLRKWVHDNVSAEVAASVRIIYGGSVNGGNCKELAAQPDVDGFLVGGASLKA-EFV 817
Query: 295 TIINSVTSKK 304
IIN+ T KK
Sbjct: 818 DIINAATVKK 847
>TC204658 homologue to UP|TPIC_ARATH (Q9SKP6) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (54%)
Length = 767
Score = 316 bits (809), Expect = 1e-86
Identities = 153/169 (90%), Positives = 164/169 (96%)
Frame = +1
Query: 139 GVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELLEEREAGKTFDVCFQQ 198
G KWV+LGHSERRH+IGE DEFIGKKAAYAL++GLGVIACIGELLEEREAGKTFDVCFQQ
Sbjct: 4 GCKWVVLGHSERRHVIGENDEFIGKKAAYALSQGLGVIACIGELLEEREAGKTFDVCFQQ 183
Query: 199 LKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRI 258
LKAYADAV SWDNIVIAYEPVWAIGTGKVA+P+QAQEVH A+RDWL+KNVS EVASKTRI
Sbjct: 184 LKAYADAVASWDNIVIAYEPVWAIGTGKVATPQQAQEVHVAVRDWLKKNVSDEVASKTRI 363
Query: 259 IYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
IYGGSVNGGNS+ELAK+EDIDGFLVGGASLKGPEFATI+NSVTSKKVAA
Sbjct: 364 IYGGSVNGGNSAELAKQEDIDGFLVGGASLKGPEFATIVNSVTSKKVAA 510
>TC204661 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (46%)
Length = 810
Score = 280 bits (717), Expect = 4e-76
Identities = 143/178 (80%), Positives = 157/178 (87%), Gaps = 3/178 (1%)
Frame = +3
Query: 2 ATSLASQLSVGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAMAGSGKFFVG 61
+TSLASQL +GLRRP KLDS NSQS SLF NLRL +S KPSR+VIAMAG+GKFFVG
Sbjct: 42 STSLASQLYIGLRRPCLKLDSFNSQS-FSLFDPNLRLSLSPPKPSRAVIAMAGTGKFFVG 218
Query: 62 GNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVS 121
GNWKCNGTKDSISKLVADLN+AKLEPDVDVVVAPPF+YIDQVKNS+T+RIE+SAQNSWV
Sbjct: 219 GNWKCNGTKDSISKLVADLNNAKLEPDVDVVVAPPFLYIDQVKNSLTERIEISAQNSWVG 398
Query: 122 KGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTE---GLGVI 176
KGGAFTGEIS EQLKD G KWV+LGHSERRH+IGE DEFIGKKAAYAL + GL +I
Sbjct: 399 KGGAFTGEISAEQLKDLGCKWVVLGHSERRHVIGENDEFIGKKAAYALIQESCGLNLI 572
>TC214994 homologue to UP|Q6GW08 (Q6GW08) Triosephosphate isomerase ,
partial (64%)
Length = 822
Score = 211 bits (536), Expect = 4e-55
Identities = 106/162 (65%), Positives = 124/162 (76%)
Frame = +2
Query: 143 VILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELLEEREAGKTFDVCFQQLKAY 202
VI ERR ++ E +EF+G K AYAL +GL VIACIGE LE+REAG T V +Q KA
Sbjct: 2 VIXXXXERRQLLNESNEFVGDKVAYALQQGLKVIACIGETLEQREAGTTTAVVSEQTKAI 181
Query: 203 ADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGG 262
A + +WDN+V+AYEPVWAIGTGKVA+P QAQEVHA +R W+ NVSAEVA+ RIIYGG
Sbjct: 182 AAKISNWDNVVLAYEPVWAIGTGKVATPAQAQEVHADLRKWVHDNVSAEVAASVRIIYGG 361
Query: 263 SVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKK 304
SVNGGN ELA + D+DGFLVGGASLK PEF IIN+ T KK
Sbjct: 362 SVNGGNCKELAAQPDVDGFLVGGASLK-PEFVDIINAATVKK 484
>TC221930 similar to UP|Q6GW08 (Q6GW08) Triosephosphate isomerase , partial
(67%)
Length = 507
Score = 144 bits (364), Expect = 4e-35
Identities = 80/169 (47%), Positives = 103/169 (60%), Gaps = 2/169 (1%)
Frame = +1
Query: 66 CNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVSKG 123
CNGT D + K+V LN AK+ E V V ++PPFV + VK+ ++ V AQN WV KG
Sbjct: 1 CNGTTDEVKKIVTTLNEAKVPGEDVVQVDMSPPFVCLTVVKSLLSPDFHVPAQNCWVRKG 180
Query: 124 GAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELL 183
+T E+S E L + G+ VI+GHSER ++ E EF+G K AYAL +GL IACI E L
Sbjct: 181 TTYTSEVSAEMLVNLGIP*VIIGHSERMQLLNESHEFLGYKVAYALQQGLKDIACIAETL 360
Query: 184 EEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQ 232
E+ EAG T V Q K+ A + N+ AYEPVWA T VA+P Q
Sbjct: 361 EQCEAGTTTAVAPDQTKSDAAK*SNCHNVGFAYEPVWAT*TRNVATPAQ 507
>BI674386
Length = 299
Score = 121 bits (304), Expect = 3e-28
Identities = 57/99 (57%), Positives = 72/99 (72%)
Frame = +2
Query: 122 KGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGE 181
KGGA+TGE+S E L + G+ WVI+GHSERR ++ E +EF+G K AYAL +GL VIACIGE
Sbjct: 2 KGGAYTGEVSAEMLVNLGIPWVIIGHSERRQLLNESNEFVGDKVAYALQQGLKVIACIGE 181
Query: 182 LLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVW 220
LE G T V +Q KA A + +WDN+V+AYEPVW
Sbjct: 182 TLELFFFGTTTAVVSEQTKAIAAKISNWDNVVLAYEPVW 298
>TC214996 UP|Q6GW08 (Q6GW08) Triosephosphate isomerase , partial (47%)
Length = 554
Score = 97.4 bits (241), Expect(2) = 9e-27
Identities = 46/79 (58%), Positives = 58/79 (73%), Gaps = 2/79 (2%)
Frame = +1
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN AK+ E V+VVV+PPFV++ VK+ + VS
Sbjct: 142 KFFVGGNWKCNGTTEEVKKIVTTLNEAKVPGEDVVEVVVSPPFVFLPFVKSLLRPDFHVS 321
Query: 115 AQNSWVSKGGAFTGEISVE 133
AQN WV KGGA+TGE+S E
Sbjct: 322 AQNCWVRKGGAYTGEVSAE 378
Score = 40.4 bits (93), Expect(2) = 9e-27
Identities = 23/59 (38%), Positives = 34/59 (56%), Gaps = 2/59 (3%)
Frame = +2
Query: 132 VEQLKDHGVKWVILGHSERRHIIGEK-DEFIGK-KAAYALTEGLGVIACIGELLEEREA 188
++ L + G+ WVI+GHSE K +EF+GK A ++ CIGE LE+RE+
Sbjct: 374 LKMLVNLGIPWVIIGHSETEAAFEMKSNEFVGK*SCLCAPARSEKLLPCIGETLEQRES 550
>AW310030 similar to SP|P48491|TPIS_ Triosephosphate isomerase cytosolic (EC
5.3.1.1) (TIM). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (38%)
Length = 295
Score = 115 bits (289), Expect = 2e-26
Identities = 60/99 (60%), Positives = 70/99 (70%)
Frame = -1
Query: 206 VPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVN 265
V +W N+V+AYEPV I TGK +P QAQEVH +R W+ NVSAEVA+ RIIY GSVN
Sbjct: 295 VSNWVNVVLAYEPVSPIATGKDRTPAQAQEVHVDLRKWVHDNVSAEVAASVRIIYAGSVN 116
Query: 266 GGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKK 304
GG L+ + D DGF VGGASLK PEF IIN+ T KK
Sbjct: 115 GGTCK*LSAQPDADGFSVGGASLK-PEFVDIINAATVKK 2
>BE800153 homologue to SP|Q9SKP6|TPIC_ Triosephosphate isomerase chloroplast
precursor (EC 5.3.1.1) (TIM). [Mouse-ear cress], partial
(17%)
Length = 338
Score = 98.2 bits (243), Expect = 4e-21
Identities = 51/56 (91%), Positives = 53/56 (94%)
Frame = -1
Query: 252 VASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
VASKTRIIYGGSVN NS+ELAK+EDIDGFLVGGASLKG EFATIINSVTSKKVAA
Sbjct: 338 VASKTRIIYGGSVNASNSAELAKQEDIDGFLVGGASLKGFEFATIINSVTSKKVAA 171
>AW781223 weakly similar to SP|P21820|TPIS Triosephosphate isomerase
cytosolic (EC 5.3.1.1) (TIM). [Japanese goldthread]
{Coptis japonica}, partial (39%)
Length = 379
Score = 92.0 bits (227), Expect = 3e-19
Identities = 57/125 (45%), Positives = 71/125 (56%), Gaps = 5/125 (4%)
Frame = +2
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKC GT D + K+V LN AK+ V VVV+P FV++ +K + VS
Sbjct: 8 KFFVGGNWKCKGTTDEVKKMVTTLNDAKVPGQNVV*VVVSPSFVFLPVIKRLLRPDYHVS 187
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIG---EKDEFIGKKAAYALTE 171
AQNSWV GGA+T + E L + + VI+GH ER I G + F YAL
Sbjct: 188 AQNSWVRIGGAYTVDS*TEMLVNLEIPLVIIGHLER--IAGY*FNQMSF*ETNVDYALQL 361
Query: 172 GLGVI 176
GL VI
Sbjct: 362 GLNVI 376
>BE190700 similar to SP|Q9SKP6|TPIC_ Triosephosphate isomerase chloroplast
precursor (EC 5.3.1.1) (TIM). [Mouse-ear cress], partial
(13%)
Length = 133
Score = 67.8 bits (164), Expect = 6e-12
Identities = 33/44 (75%), Positives = 39/44 (88%)
Frame = +2
Query: 264 VNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
VN N +ELAK+EDI+G++VGGA LKGPEFATIINS+TSKK AA
Sbjct: 2 VNASNRAELAKQEDIEGWIVGGA*LKGPEFATIINSITSKKGAA 133
>BE440904 similar to SP|P21820|TPIS_ Triosephosphate isomerase cytosolic (EC
5.3.1.1) (TIM). [Japanese goldthread] {Coptis japonica},
partial (21%)
Length = 352
Score = 66.6 bits (161), Expect = 1e-11
Identities = 42/116 (36%), Positives = 60/116 (51%), Gaps = 3/116 (2%)
Frame = +2
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + + LN AK+ E ++VVV+PPFV + K+ + +
Sbjct: 8 KFFVGGNWKCNGTTEEVKNIATTLNDAKVPGEDVLEVVVSPPFVRLPVGKSLLLPYLHDM 187
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILG-HSERRHIIGEKDEFIGKKAAYAL 169
Q+ GGA+ S L + V+ G H E R II + + G+ Y L
Sbjct: 188 EQSC*FRNGGAYLVVASAAMLVKLPISCVVSGIHHEGRIIINDV-SWTGEMGVYEL 352
>BU765692 similar to SP|Q9M4S8|TPIC_ Triosephosphate isomerase chloroplast
precursor (EC 5.3.1.1) (TIM). [Strawberry], partial
(7%)
Length = 339
Score = 46.6 bits (109), Expect = 1e-05
Identities = 22/23 (95%), Positives = 23/23 (99%)
Frame = +2
Query: 67 NGTKDSISKLVADLNSAKLEPDV 89
NGTKDSISKLVADLN+AKLEPDV
Sbjct: 26 NGTKDSISKLVADLNNAKLEPDV 94
>AI966765 weakly similar to SP|P21820|TPIS_ Triosephosphate isomerase
cytosolic (EC 5.3.1.1) (TIM). [Japanese goldthread]
{Coptis japonica}, partial (13%)
Length = 106
Score = 37.4 bits (85), Expect = 0.009
Identities = 19/36 (52%), Positives = 25/36 (68%)
Frame = +2
Query: 268 NSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSK 303
NS+ A++ D DGF VGGASLK P+F I + T+K
Sbjct: 2 NSNNWAEQPDADGFWVGGASLK-PDFVDTITAATAK 106
>TC214290 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp90-2, complete
Length = 2566
Score = 32.7 bits (73), Expect = 0.21
Identities = 22/95 (23%), Positives = 44/95 (46%)
Frame = +1
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K +D ++ ++ + F +E+LK G + + + + + +G+
Sbjct: 1561 YVTRMKEGQSDIYYITGESKKAVENSPF-----LEKLKKKGYEVLFMVDAIDEYAVGQLK 1725
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + FD
Sbjct: 1726 EFEGKKLVSATKEGLKLDESEDEKKKQEELKEKFD 1830
>TC214192 similar to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp90-2, partial
(48%)
Length = 1223
Score = 32.3 bits (72), Expect = 0.28
Identities = 22/95 (23%), Positives = 44/95 (46%)
Frame = +2
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K +D ++ ++ + F +E+LK G + + + + + +G+
Sbjct: 272 YVTRMKEGQSDIYYITGESKKAVENSPF-----LEKLKKKGYEVLYMVDAIDEYAVGQLK 436
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + FD
Sbjct: 437 EFEGKKLVSATKEGLKLDESEDEKKKQEELKEKFD 541
>TC224706 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp90-2, complete
Length = 2385
Score = 30.0 bits (66), Expect = 1.4
Identities = 21/95 (22%), Positives = 43/95 (45%)
Frame = +3
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K D ++ ++ + F +E+LK G + + + + + +G+
Sbjct: 1404 YVTRMKEGQNDIYYITGESKKAVENSPF-----LEKLKKKGFEVLYMVDAIDEYAVGQLK 1568
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + F+
Sbjct: 1569 EFEGKKLVSATKEGLKLDESEDEKKKKEELKEKFE 1673
>TC215641 homologue to UP|O04091 (O04091) Endomembrane protein EMP70 precusor
isolog; 68664-64364, partial (11%)
Length = 658
Score = 28.9 bits (63), Expect = 3.1
Identities = 14/47 (29%), Positives = 22/47 (46%)
Frame = +3
Query: 42 SSKPSRSVIAMAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPD 88
S +P + +AG KF G W C GTK + V + +++ D
Sbjct: 210 SLRPEHFHMFVAGDNKFLRGSLWSCVGTKQCLQ--VVSITGLRMDGD 344
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,438,630
Number of Sequences: 63676
Number of extensions: 159565
Number of successful extensions: 685
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 677
length of query: 307
length of database: 12,639,632
effective HSP length: 97
effective length of query: 210
effective length of database: 6,463,060
effective search space: 1357242600
effective search space used: 1357242600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0012b.1


