

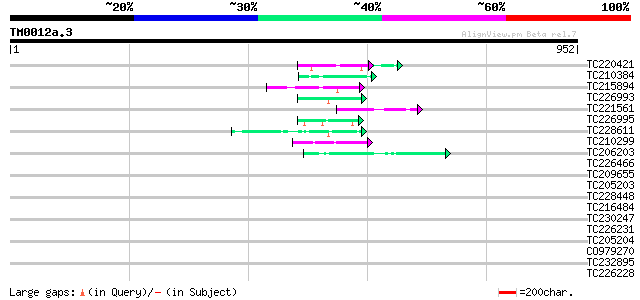
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.3
(952 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 46 9e-05
TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_6... 45 2e-04
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 45 2e-04
TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 deh... 44 3e-04
TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 44 3e-04
TC226995 similar to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehyd... 44 3e-04
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 43 6e-04
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 43 8e-04
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 43 8e-04
TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {... 42 0.001
TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 42 0.001
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 42 0.002
TC228448 weakly similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (4%) 41 0.003
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 40 0.004
TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g6... 40 0.005
TC226231 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement p... 40 0.005
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 40 0.006
CO979270 40 0.006
TC232895 weakly similar to PRF|1604369A.0|226743|1604369A sulfat... 40 0.006
TC226228 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement p... 40 0.006
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 45.8 bits (107), Expect = 9e-05
Identities = 53/184 (28%), Positives = 70/184 (37%), Gaps = 7/184 (3%)
Frame = +3
Query: 483 HSSNPLTPIPESQPAEQTTSPPH-----SPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTS 537
H +PL + QP +TSPPH SP S+ P+ P + PS S PT+
Sbjct: 156 HHPHPLWTQNKQQPLNPSTSPPHRTPAPSPPSTM---PPSATPPSPSATSSPSAS--PTA 320
Query: 538 LLTIPYDPLSSEPIIHEQPEPNQTEPQ-PRTSDHSAPRASARPAARTTDTDSSTAFTPVS 596
T P S P P P+ T P P S ++P S A T +S + +P S
Sbjct: 321 PPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTS 500
Query: 597 FPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPD-EPI 655
+SPS S S K A PS + P P P + PD P
Sbjct: 501 ---RTSPTSPSPTSTS----------KPPAPSSSSPA*PSSKPSPSPTP---ISPDPSPA 632
Query: 656 LANP 659
+ P
Sbjct: 633 TSTP 644
Score = 44.7 bits (104), Expect = 2e-04
Identities = 38/135 (28%), Positives = 62/135 (45%), Gaps = 8/135 (5%)
Frame = +3
Query: 484 SSNPLTPIPESQPAEQTTSPPHSPRSSF-FQPSPTEAPLWNLLQNQPSRSDEPTSLLTIP 542
S+ P +P S P+ T+PP SP S PSP AP + P + PTS P
Sbjct: 264 SATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSP 443
Query: 543 YDPLSSEPIIHEQPEPNQTEPQPRTSDHS-APRASARPAARTTDTDS------STAFTPV 595
P S+ P P RTS S +P ++++P A ++ + + S + TP+
Sbjct: 444 PSPASTA----SANSPASGSPTSRTSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPI 611
Query: 596 SFPINVVDSSPSNNS 610
S + S+P++++
Sbjct: 612 SPDPSPATSTPTSHT 656
Score = 37.7 bits (86), Expect = 0.024
Identities = 49/181 (27%), Positives = 75/181 (41%), Gaps = 4/181 (2%)
Frame = +3
Query: 420 QKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISA 479
Q K++ + P PA T PP+ S + +S P+ + A P S
Sbjct: 180 QNKQQPLNPSTSPPHRTPAPSPPS-TMPPSATPPSPSATSSPSASPT-------APPTSP 335
Query: 480 LLQHSSNPLTPIPESQPAEQTT--SPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTS 537
S P +P P S P+ +T SPP + + PSP +P N P+ S PTS
Sbjct: 336 ---SPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSP-PSPASTASANSPA-SGSPTS 500
Query: 538 LLTIPYDPLSSEPIIHEQPEPNQTEPQ-PRTSDHSAPR-ASARPAARTTDTDSSTAFTPV 595
T P P S + P P+ + P P + +P S P+ T+ S T+ +P+
Sbjct: 501 -RTSPTSP--SPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISPI 671
Query: 596 S 596
+
Sbjct: 672 T 674
>TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_60, partial
(11%)
Length = 860
Score = 44.7 bits (104), Expect = 2e-04
Identities = 44/134 (32%), Positives = 54/134 (39%), Gaps = 2/134 (1%)
Frame = +3
Query: 485 SNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
S L P S P +T +P SP +S F S P NQPS+ TS
Sbjct: 333 SPSLPPTTASLPTRRTPTP--SPTTSSFASSKPPPP-----SNQPSKPSSKTSSSKTKNI 491
Query: 545 PLSSEPIIHEQ-PEPNQTEPQPRTSDHSAPRASARPAARTTDTDSST-AFTPVSFPINVV 602
S P P P+ TS +AP ASA PA + T S T A TP S P +
Sbjct: 492 NF*SSPTSSSAGPPPSPRNSASSTSSSAAPPASASPATTPSGTISPTAASTPTSSPYRI- 668
Query: 603 DSSPSNNSESIRKF 616
SP S + R +
Sbjct: 669 --SPKRVSFTARSY 704
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 44.7 bits (104), Expect = 2e-04
Identities = 45/171 (26%), Positives = 69/171 (40%), Gaps = 6/171 (3%)
Frame = +1
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNP--LT 489
P+ E R R + +S+ SS P TS +A SS P +
Sbjct: 280 PSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPTSS-----------AARCSTSSTPPPSS 426
Query: 490 PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS- 548
P P S P SPP +P S+ P+P +P + S S TS + P PLSS
Sbjct: 427 PAPRSTPPPTPPSPP-APTSASSPPAPARSP-----ASHASTSSRGTSPSSAPLFPLSSV 588
Query: 549 ---EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVS 596
+ P P+ + P R S ++P ++ A T +S + +P++
Sbjct: 589 TPPTQLSSSFPTPSTSSPTSRGSSPASPPTASSAPAPTWTLPASVSSSPIT 741
Score = 38.5 bits (88), Expect = 0.014
Identities = 36/126 (28%), Positives = 49/126 (38%), Gaps = 2/126 (1%)
Frame = +1
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPL-WNLLQNQPSR-SDEPTSLLTIPYDPL 546
+P P S P T+PP PR S SP+ AP W +PS PTS + P
Sbjct: 208 SPRPSSGP*---TTPPLPPRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPT 378
Query: 547 SSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVVDSSP 606
SS P + P PR++ P + P T +S+ P P + +S
Sbjct: 379 SSAARCSTSSTPPPSSPAPRSTPPPTPPSPPAP------TSASSPPAPARSPASHASTSS 540
Query: 607 SNNSES 612
S S
Sbjct: 541 RGTSPS 558
Score = 35.4 bits (80), Expect = 0.12
Identities = 51/209 (24%), Positives = 76/209 (35%), Gaps = 3/209 (1%)
Frame = +1
Query: 561 TEPQPRTSDHSAPRASARPAARTTDTDSSTAF--TPVSFPINVVDSSPSNNSESIRKFME 618
T P+P + + P RP+A T S+ A P P ++ S S++S +
Sbjct: 205 TSPRPSSGP*TTPPLPPRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPTSS 384
Query: 619 VRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILA-NPIQEADPLVQQAHPVPDQQ 677
+ S+ + P+PR P P P P P A +P A A
Sbjct: 385 AARCSTSSTPP--PSSPAPRSTPPPTPP---SPPAPTSASSPPAPARSPASHASTSSRGT 549
Query: 678 EPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHP 737
P P SV+ + + S P TS P TS ++P SP AS A
Sbjct: 550 SPSSAPLFPLSSVTPPTQLSSSFPTPSTSSP---TSRGSSP----ASPPTASSA------ 690
Query: 738 ASPAPNLSIIPYTHLQPTSLSECINIFYH 766
PAP ++ P +L+ + H
Sbjct: 691 --PAPTWTLPASVSSSPITLTSTPRTYRH 771
>TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (94%)
Length = 1388
Score = 44.3 bits (103), Expect = 3e-04
Identities = 38/132 (28%), Positives = 53/132 (39%), Gaps = 17/132 (12%)
Frame = +2
Query: 484 SSNPLTPIPESQPAEQT-TSPPHSPRS--SFFQPSPTEAPLWNLLQNQPSRSDE------ 534
S+ P++P P A T +SP S S S PSP+ P + P+ S
Sbjct: 314 STTPISPTPPPSAAGSTPSSPTRSTTSPLSLTSPSPSRYPTTPPTSSPPAPSASSRPSVP 493
Query: 535 -------PTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASAR-PAARTTDT 586
PTS T P P S P + P+P + P P T +AP AR T +
Sbjct: 494 TSPPPAAPTSATTKPAPPRCSAPPLRRSPKPPPSTPAPPTPPPNAPPTGTP*TTARLTPS 673
Query: 587 DSSTAFTPVSFP 598
+TA + + P
Sbjct: 674 SPATASSSTTSP 709
Score = 34.7 bits (78), Expect = 0.20
Identities = 51/178 (28%), Positives = 61/178 (33%), Gaps = 1/178 (0%)
Frame = +2
Query: 565 PRTSDHSAPRASAR-PAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEK 623
PRTS SA S P T SST + P + S+PS+ + S +
Sbjct: 239 PRTSTRSASTTSMWIPTTPTRPA*SSTTPISPTPPPSAAGSTPSSPTRSTTSPLS----- 403
Query: 624 VSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPD 683
LT PSP RYP P P + P P A P +P P
Sbjct: 404 --------LTSPSPSRYP-TTPPTSSPPAPSASSRPSVPTSP--PPAAPTSATTKPAPP- 547
Query: 684 PEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPA 741
S RSP P T P T PNAP P G + P+SPA
Sbjct: 548 -----RCSAPPLRRSPKPPPSTPAP--PTPPPNAP------PTGTP*TTARLTPSSPA 682
>TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate dehydrogenase,
complete
Length = 2001
Score = 44.3 bits (103), Expect = 3e-04
Identities = 40/148 (27%), Positives = 65/148 (43%), Gaps = 4/148 (2%)
Frame = -1
Query: 549 EPIIHEQPEPNQTEPQPRT-SDHS--APRASARPAARTTDTDSSTAFTPVSFPINVVDSS 605
+P H QP+P +++ QP T S HS + +A +P++ T+ + + +P ++P +
Sbjct: 1122 QPSTH-QPQP-ESDSQPGT*SQHSPQSHQAQGKPSSHTSPSLTDKLSSPENYPTLQHSTQ 949
Query: 606 PSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYP-GPRPERLVDPDEPILANPIQEAD 664
PS N+ + Y+L PR P PRP P P+ NP
Sbjct: 948 PSPNAPT---------------SPYHLPSSPPRPPP*TPRPPP-PSPAPPVSGNPSPSPP 817
Query: 665 PLVQQAHPVPDQQEPIQPDPEPEQSVSN 692
P P P Q+P P P+P+ S+
Sbjct: 816 P------PSPPNQQPHHPSPKPQHHFSH 751
>TC226995 similar to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (59%)
Length = 679
Score = 43.9 bits (102), Expect = 3e-04
Identities = 40/134 (29%), Positives = 52/134 (37%), Gaps = 23/134 (17%)
Frame = +3
Query: 484 SSNPLTPIPE--------SQPAEQTTSPPHSPRSSFFQPSPTEAPLWN----------LL 525
S+ P++P P S P TTSPP SP S SPT P + +
Sbjct: 213 STTPISPTPPPSAAGSTPSSPTRSTTSPP-SPTSPTHSRSPTTPPTSSPPAPSAYSRPIA 389
Query: 526 QNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPR-----ASARPA 580
P+ + PTS T P P S P + +P P P T +AP +ARP
Sbjct: 390 PTSPTTA-APTSATTKPAPPRCSAPPLRRNRKPLACTPAPHTPAANAPTTGTP*TTARPT 566
Query: 581 ARTTDTDSSTAFTP 594
+ T SS P
Sbjct: 567 PSSPATASSLTNDP 608
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 43.1 bits (100), Expect = 6e-04
Identities = 59/233 (25%), Positives = 79/233 (33%), Gaps = 7/233 (3%)
Frame = +3
Query: 373 FPSPPKRKAQRKMVLDESSEESDVPLVKKTKRK---PDDGDDDDSEDGPPQKKKKKVRIV 429
F +PP R +S S LV+ + R P + PP +
Sbjct: 66 FSAPPTR---------DSPWTSTRTLVRSSARS*GTPSQASRSPAPPPPPPRSASSSTTA 218
Query: 430 VKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLT 489
PT V P + R P T +S S PA S SS P
Sbjct: 219 SSPTAVTPPSSSPPRPSPGPSATPTST-SPSPATPSTS----------------SSAPRP 347
Query: 490 PIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSD----EPTSLLTIPYDP 545
P+ S PA P SP +F P P + + + PS S EP S PY P
Sbjct: 348 PL--SSPA-----PTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEPPS--PPPYPP 500
Query: 546 LSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFP 598
S P PN + S +S+P P+ T SS+ +P + P
Sbjct: 501 TSPPPPCPSLKSPNSSPNAASPSKNSSPSVGPTPSDSLT-APSSSRTSPTTLP 656
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 42.7 bits (99), Expect = 8e-04
Identities = 34/136 (25%), Positives = 62/136 (45%), Gaps = 2/136 (1%)
Frame = +2
Query: 476 PISALLQHSSNP--LTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSD 533
PI + L+ ++ P +P+P S P + +TSP +P + PSP+ P + P+ +
Sbjct: 80 PIISSLKPTNPPPHTSPLPSSTPPQSSTSPLPTPPPA-AAPSPSAPPAASSSARSPTSTS 256
Query: 534 EPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFT 593
P++ T P S P P+ P T+ + P SA A+ +T S+T
Sbjct: 257 APSATSTTVRPP--SPPPSPWPSPPSAIVPLKNTTRSTPPPRSAPAASPSTPPPSNTRPR 430
Query: 594 PVSFPINVVDSSPSNN 609
+ P + ++P+ +
Sbjct: 431 TATTPTSTAPATPTTS 478
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 42.7 bits (99), Expect = 8e-04
Identities = 67/255 (26%), Positives = 88/255 (34%), Gaps = 8/255 (3%)
Frame = +1
Query: 494 SQPAEQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPII 552
S P TSP +SP S + P SPT +P + P S PTS P P S
Sbjct: 1138 SSPGYSPTSPGYSPTSPGYSPTSPTYSP------SSPGYS--PTSPAYSPTSPSYSPTSP 1293
Query: 553 HEQP-EPNQTEPQPRTSDHSAPRASARPAARTTD---TDSSTAFTPVSFPINVVDSSPSN 608
P P+ + P S S + PA T + +S A++P S + S S
Sbjct: 1294 SYSPTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSP 1473
Query: 609 NSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPILANPIQEADPLVQ 668
S S Y T PS Y P P P + P
Sbjct: 1474 TSPS-----------------YSPTSPS---YSPTSPS--YSPTSPAYSPTSPGYSPTSP 1587
Query: 669 QAHPVPDQQEPIQPDPEPEQSVSNQSSVRSP-HPLVETSDPHLGTSEPNAPMINIGSPQG 727
P P P P+ + + S SP P + S P+ TS +P SP
Sbjct: 1588 SYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTS 1767
Query: 728 ASEAHSS--NHPASP 740
S + SS P+SP
Sbjct: 1768 PSYSPSSPTYSPSSP 1812
Score = 35.4 bits (80), Expect = 0.12
Identities = 54/233 (23%), Positives = 79/233 (33%), Gaps = 9/233 (3%)
Frame = +1
Query: 476 PISALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNQPSRSDE 534
P S SS +P + PA TSP +SP S + P SP+ +P + + S S
Sbjct: 1198 PTSPTYSPSSPGYSP---TSPAYSPTSPSYSPTSPSYSPTSPSYSPT-SPSYSPTSPSYS 1365
Query: 535 PTSLLTIPYDPLSSEPIIHEQP-EPNQTEPQPRTSDHSAPRASARPAARTTD---TDSST 590
PTS P P S P P+ + P S S + P+ T + +S
Sbjct: 1366 PTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSP 1545
Query: 591 AFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGPRPERLVD 650
A++P S + S S S S + + PS R P
Sbjct: 1546 AYSPTSPGYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTS 1725
Query: 651 PDEPILANPIQEADPLVQQAHPVPDQQEP----IQPDPEPEQSVSNQSSVRSP 699
P+ + P + P P + PD P + S+ SP
Sbjct: 1726 PNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFSPSTGYSP 1884
Score = 29.3 bits (64), Expect = 8.6
Identities = 24/92 (26%), Positives = 37/92 (40%), Gaps = 1/92 (1%)
Frame = +1
Query: 486 NPLTPIPESQPAEQTTSPPHSPRSSFFQP-SPTEAPLWNLLQNQPSRSDEPTSLLTIPYD 544
+P +P + P TSP +SP S + P SPT +P S S ++ Y
Sbjct: 1696 SPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSP-----------SSPYNSGVSPDYS 1842
Query: 545 PLSSEPIIHEQPEPNQTEPQPRTSDHSAPRAS 576
P S + P+Q P ++ P+ S
Sbjct: 1843 PSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTS 1938
>TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {Arabidopsis
thaliana;} , partial (22%)
Length = 978
Score = 42.4 bits (98), Expect = 0.001
Identities = 49/203 (24%), Positives = 78/203 (38%), Gaps = 9/203 (4%)
Frame = +1
Query: 418 PPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPI 477
PP + + KP V P A V P T + P ++ L LP
Sbjct: 175 PPTISQPPAIVAPKPAPVTPPAPKVAPASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLPP 354
Query: 478 SALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSF-------FQPSPTEAPLWNLLQNQPS 530
+ ++ P P P+ P TSP +P + +P+PT AP+ P
Sbjct: 355 PPV---ATPPPLPPPKIAPTPAKTSPAPAPAKATPAPAPAPTKPAPTPAPI------PPP 507
Query: 531 RSDEPTSLLTIPYD-PLSSEPIIHEQPEPNQTEPQPR-TSDHSAPRASARPAARTTDTDS 588
+ PT ++ +P P S + H P P T H +P A P T ++D+
Sbjct: 508 PTLAPTPIVEVPAPAPSSHKHRRHRHRHKRHQAPAPAPTVIHKSPPA---PPDSTAESDT 678
Query: 589 STAFTPVSFPINVVDSSPSNNSE 611
TP P ++++PSN+ +
Sbjct: 679 ----TPAPSPSLNLNAAPSNHQQ 735
Score = 35.4 bits (80), Expect = 0.12
Identities = 49/241 (20%), Positives = 80/241 (32%), Gaps = 3/241 (1%)
Frame = +1
Query: 516 PTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRA 575
P +P L PS S P ++ P +S P QP P P+P AP+
Sbjct: 76 PANSPPTTLAATSPS-SGTPPAVTQAASPPTTSNPPTISQP-PAIVAPKPAPVTPPAPKV 249
Query: 576 SARPAARTTDTDSSTAFTPVSFPINVVDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCP 635
+ + + + P P+ P + + K++ P
Sbjct: 250 APASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLPPPPVATPPPLPPPKIAPTPAKTSPAP 429
Query: 636 SPRRY---PGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSN 692
+P + P P P + PI P P+V+ P P + +
Sbjct: 430 APAKATPAPAPAPTKPAPTPAPIPPPPTLAPTPIVEVPAPAPSSH-------KHRRHRHR 588
Query: 693 QSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHL 752
++P P P + P AP P +E+ ++ P SP+ NL+ P H
Sbjct: 589 HKRHQAPAPA-----PTVIHKSPPAP------PDSTAESDTTPAP-SPSLNLNAAPSNHQ 732
Query: 753 Q 753
Q
Sbjct: 733 Q 735
>TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (32%)
Length = 832
Score = 42.4 bits (98), Expect = 0.001
Identities = 40/132 (30%), Positives = 57/132 (42%), Gaps = 6/132 (4%)
Frame = +1
Query: 487 PLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQN--QP-SRSDEPTSLLTIPY 543
P T P + A TTSPP SP S +P PT P L + QP S S PT+ +
Sbjct: 34 PTTSSPSTTAA--TTSPPSSPSSPTHRPLPTRTPTQPLPPHSAQPSSNSPSPTAPSRLHS 207
Query: 544 DPLSSEPIIHEQPEPNQTEPQ---PRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPIN 600
SS P P P R + ++P +S+ P++ T ++ P S+ +
Sbjct: 208 GTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAAPKSSTKP-SYSPS 384
Query: 601 VVDSSPSNNSES 612
V S PS + S
Sbjct: 385 VRSSLPSPTTRS 420
Score = 34.7 bits (78), Expect = 0.20
Identities = 45/192 (23%), Positives = 69/192 (35%), Gaps = 12/192 (6%)
Frame = +1
Query: 418 PPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPI 477
P Q KK PT P+ T PP+ + S H P T P
Sbjct: 7 PNQSSSKKA-----PTTSSPSTTAAT-TSPPSSPS-SPTHRPLPTRT-----------PT 132
Query: 478 SALLQHSSNPL--TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLW----NLLQNQPSR 531
L HS+ P +P P + + + SP + PSP+ P + N + P+
Sbjct: 133 QPLPPHSAQPSSNSPSPTAPSRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTS 312
Query: 532 SDEPTSLLTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASAR------PAARTTD 585
S P+S S++P + P R+S + A++R P T
Sbjct: 313 SSTPSSSTPSAAPKSSTKPSYSPSVRSSLPSPTTRSSARAPETATSRKP*T*CPKCAATV 492
Query: 586 TDSSTAFTPVSF 597
T +++ T SF
Sbjct: 493 TSRTSSTTAPSF 528
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 41.6 bits (96), Expect = 0.002
Identities = 30/119 (25%), Positives = 45/119 (37%), Gaps = 2/119 (1%)
Frame = +3
Query: 484 SSNPLTPIPESQPAEQTTSPPHSPRS--SFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTI 541
S++P P P++ P +SPP P S P P P + L + P S P+S
Sbjct: 279 STSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPS 458
Query: 542 PYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPIN 600
P S P P + P P + +P + P A + TP P++
Sbjct: 459 SPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLS 635
Score = 41.2 bits (95), Expect = 0.002
Identities = 40/147 (27%), Positives = 59/147 (39%), Gaps = 7/147 (4%)
Frame = +3
Query: 437 PAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQP 496
PA + + PP + + SS P S + P+S+ S P +P P S P
Sbjct: 288 PAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPP 467
Query: 497 AEQ---TTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSL----LTIPYDPLSSE 549
++ PP SP S P P+ P ++ P + P ++ LT PLSS
Sbjct: 468 PASPPPSSPPPASPPPS--SPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSP 641
Query: 550 PIIHEQPEPNQTEPQPRTSDHSAPRAS 576
P+ P P++ P S AP S
Sbjct: 642 PVSSPAPSPSKVF-APALSPSLAPGPS 719
Score = 33.1 bits (74), Expect = 0.60
Identities = 33/125 (26%), Positives = 44/125 (34%), Gaps = 1/125 (0%)
Frame = +3
Query: 635 PSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDP-EPEQSVSNQ 693
P P P P P P P ++P + P A P P P P P P S
Sbjct: 375 PPPASTPPPSPLSSPPPSSPPPSSPPPSSPP---PASPPPSSPPPASPPPSSPPPSSPPP 545
Query: 694 SSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQ 753
S P T P + + + SP +S A S + +PA + S+ P L
Sbjct: 546 FSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVSSPAPSPSKVFAPALSPSLAPGPSLS 725
Query: 754 PTSLS 758
S S
Sbjct: 726 TISPS 740
>TC228448 weakly similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (4%)
Length = 458
Score = 40.8 bits (94), Expect = 0.003
Identities = 39/120 (32%), Positives = 55/120 (45%), Gaps = 7/120 (5%)
Frame = +1
Query: 484 SSNPLTPIPE-----SQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSL 538
S +PL P P + P+ T +PP SP S +PSP N P P L
Sbjct: 109 SPSPLKP*PYPHSNLASPSGPTPNPPFSPTS---KPSPAPRSSPPSRSNSPPPHVPPLRL 279
Query: 539 LTIPYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRAS-ARPAARTT-DTDSSTAFTPVS 596
L+ P +S P ++P PR S S+P +S + +ARTT + SS+A +P S
Sbjct: 280 LSKPTTASTSRS--PSPLSPTSSKPTPRPSPRSSPSSSRSSSSARTTRRSPSSSASSPRS 453
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 40.4 bits (93), Expect = 0.004
Identities = 50/201 (24%), Positives = 69/201 (33%), Gaps = 16/201 (7%)
Frame = +1
Query: 400 KKTKRKPDDGDDDDSEDGPPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSS 459
+ T P S PP+ K T P R+ PP+ +R+S+ S+
Sbjct: 133 QSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTT--PPCRTFSRSTPPSTPSRTSSPST 306
Query: 460 KPALTSDDDLNLFDALPISALLQHSSNPLTP------IPESQPAEQTTSPPHSPRSSFFQ 513
+ TS P S+ ++ P P P P +TSP + S
Sbjct: 307 SSSTTS---------APRSSTRSPTAPPSPPQCTKPPAPPPDPPASSTSPTSTAEKSHSP 459
Query: 514 PSPTEAPLWNLLQNQPSRSDEP--TSLLT------IPYDPLSSEPIIHEQPEPNQTEPQ- 564
P T A + P RS P SL T + PL +H P PN+T P
Sbjct: 460 PKTTTA-------HSPQRSSNP*KKSLTTSQ*SRSAKFSPLPQRKPLHPHP-PNKTSPTL 615
Query: 565 -PRTSDHSAPRASARPAARTT 584
T S+P S R T
Sbjct: 616 CQNTDARSSPTLSLLNPTRLT 678
Score = 30.0 bits (66), Expect = 5.0
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = +2
Query: 368 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDDDSED 416
P A + P+P K ++K D +++SD P PDD D ++D
Sbjct: 1067 PSAADAPAPAKNGKKKKKAADAPADDSDAP-----ADSPDDAADVTADD 1198
>TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61870
{Arabidopsis thaliana;} , partial (56%)
Length = 1291
Score = 40.0 bits (92), Expect = 0.005
Identities = 38/122 (31%), Positives = 55/122 (44%), Gaps = 3/122 (2%)
Frame = +1
Query: 489 TPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSS 548
TP P S P T PP P+S T + +P+RS TS + P PL+
Sbjct: 55 TPPPPSSPR---THPPP*PQSKKPAAPSTSS------NRRPTRSASSTSAVPPP-SPLTL 204
Query: 549 EPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSS--TAFTPVSFPI-NVVDSS 605
P P+ + P P TS SAP ++ A T+ T +S T + + PI ++ S+
Sbjct: 205 TSTAVPSPLPSPSSPPPTTSPASAPSSTISKPAPTSATKNSSPTPSSSTARPICSITPSA 384
Query: 606 PS 607
PS
Sbjct: 385 PS 390
Score = 36.6 bits (83), Expect = 0.054
Identities = 41/164 (25%), Positives = 61/164 (37%), Gaps = 2/164 (1%)
Frame = +1
Query: 418 PPQKKKKKVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPI 477
PP + P +PAA P R +S+ S+ P P
Sbjct: 58 PPPPSSPRTHPPP*PQSKKPAAPSTSSNRRPTR--SASSTSAVP--------------PP 189
Query: 478 SALLQHSSNPLTPIPESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTS 537
S L S+ +P+P TTSP +P S+ +P+PT A + S + S
Sbjct: 190 SPLTLTSTAVPSPLPSPSSPPPTTSPASAPSSTISKPAPTSATKNSSPTPSSSTARPICS 369
Query: 538 LLTIPYDPLSSEPIIHEQP-EPNQTEP-QPRTSDHSAPRASARP 579
+ P +S P+ +P P+ P PRT+ S S P
Sbjct: 370 ITPSAPSPKTSPPLAPSKP*IPSSLLPSSPRTTKSSLESTSNSP 501
>TC226231 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement pol
polyprotein, partial (97%)
Length = 1375
Score = 40.0 bits (92), Expect = 0.005
Identities = 36/149 (24%), Positives = 63/149 (42%), Gaps = 10/149 (6%)
Frame = +1
Query: 802 QVPIMMQLLHAEGS----QSIEAAKQRFARRVALHE---LEQRNKLLEAIEEAKRKKEQA 854
Q+ I Q++ EG+ ++++A + A A+ + ++ +K ++ I E +Q
Sbjct: 397 QLRIHDQMIMLEGAKATTETVDALRTGAAAMRAMQKETNIDDVDKTMDEINEQTENMKQI 576
Query: 855 EEAARQ---AAAQAEQARLEAERLEAEAEALRCQALAPVVFTPAASASIPDPQAAQNVPS 911
+EA AAA ++ LEAE E E L Q L P PAA +P S
Sbjct: 577 QEALSTPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAAPVHVPAGWQPTRPVS 756
Query: 912 SSTQTSSSRLDIMEQRLDTHESMLIEMKH 940
S L ++ + S +++H
Sbjct: 757 SKPTAEEDELAALQAEMAL*ASPFSQLRH 843
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 39.7 bits (91), Expect = 0.006
Identities = 40/162 (24%), Positives = 56/162 (33%), Gaps = 2/162 (1%)
Frame = +1
Query: 484 SSNPLTPIPESQPAEQTTSPPHSPRS--SFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTI 541
S++P P P+S P +SPP P S P P P L + P S P+S
Sbjct: 187 STSPAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASPPPSS---- 354
Query: 542 PYDPLSSEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINV 601
P S P P P P + ++P + P A A P + P
Sbjct: 355 -PPPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASPPPFSPPPATPPPATPPPA 531
Query: 602 VDSSPSNNSESIRKFMEVRKEKVSALEEYYLTCPSPRRYPGP 643
+ +P ++ + K AL SP PGP
Sbjct: 532 LTPTPLSSPPATSPAPAPAKVVAPAL--------SPSLAPGP 633
Score = 35.4 bits (80), Expect = 0.12
Identities = 41/166 (24%), Positives = 53/166 (31%)
Frame = +1
Query: 437 PAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPESQP 496
PA + + PP + + SS P S + P+S+ S P +P P S P
Sbjct: 196 PAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASPP 375
Query: 497 AEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHEQP 556
+ PP SP + P P P P S P
Sbjct: 376 --PASPPPASPPPA--SPPPASPP------------------------PASPPPATPPPA 471
Query: 557 EPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTPVSFPINVV 602
P P P T P A+ PA T S A +P P VV
Sbjct: 472 SPPPFSPPPAT----PPPATPPPALTPTPLSSPPATSPAPAPAKVV 597
Score = 30.4 bits (67), Expect = 3.9
Identities = 30/125 (24%), Positives = 39/125 (31%), Gaps = 1/125 (0%)
Frame = +1
Query: 635 PSPRRYPGPRPERLVDPDEPILANPIQEADPLVQQAHPVPDQQEPIQPDPEPEQSVSNQS 694
P P P P P P P ++P + P P P P P
Sbjct: 283 PPPASTPPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATP 462
Query: 695 SVRSPHPL-VETSDPHLGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLSIIPYTHLQ 753
SP P + P T P + SP S A + +PA + S+ P L
Sbjct: 463 PPASPPPFSPPPATPPPATPPPALTPTPLSSPPATSPAPAPAKVVAPALSPSLAPGPSLS 642
Query: 754 PTSLS 758
S S
Sbjct: 643 TISPS 657
Score = 30.4 bits (67), Expect = 3.9
Identities = 27/86 (31%), Positives = 31/86 (35%), Gaps = 2/86 (2%)
Frame = +1
Query: 659 PIQEADPLVQQAHPVPDQQEP--IQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPN 716
P P Q+ P P Q P + P P QS +S P PL S P
Sbjct: 184 PSTSPAPPTPQSSPPPIQSSPPPVPSSPPPAQSPP-PASTPPPAPLSSPPPASPPPSSPP 360
Query: 717 APMINIGSPQGASEAHSSNHPASPAP 742
SP AS +S PASP P
Sbjct: 361 PASPPPASPPPASPPPASPPPASPPP 438
>CO979270
Length = 837
Score = 39.7 bits (91), Expect = 0.006
Identities = 41/156 (26%), Positives = 58/156 (36%), Gaps = 12/156 (7%)
Frame = +3
Query: 436 EPAAEVVRR-TEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPIPES 494
E A V ++ T P T+ S + L FD+ SA L S L +P
Sbjct: 258 EQAKRVTKQGTSYPTPATKGSTNEQHSQLLFRPCGKFFDSPGASAALTMPSQKLLSLPHL 437
Query: 495 QPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLSSEPIIHE 554
++P H+P PSP+ ++L+ +P P P S E
Sbjct: 438 SHLTF*SNPEHTP----IPPSPSSPSRCSILRRKP------------PSPPQS------E 551
Query: 555 QPEPNQTEPQ-----------PRTSDHSAPRASARP 579
QPEP+ + PQ P DH P A+A P
Sbjct: 552 QPEPHNSAPQQASEQNLHPQEPHQLDHFQPNAAASP 659
Score = 29.3 bits (64), Expect = 8.6
Identities = 33/121 (27%), Positives = 52/121 (42%), Gaps = 6/121 (4%)
Frame = +3
Query: 632 LTCPSPRRYPGPRPERLV---DPDE-PILANPIQEAD-PLVQQAHPVPDQQEPIQPDPEP 686
LT PS + P L +P+ PI +P + ++++ P P Q E PEP
Sbjct: 396 LTMPSQKLLSLPHLSHLTF*SNPEHTPIPPSPSSPSRCSILRRKPPSPPQSE----QPEP 563
Query: 687 EQSVSNQSSVRSPHPLVETSDPH-LGTSEPNAPMINIGSPQGASEAHSSNHPASPAPNLS 745
S Q+S ++ HP +PH L +PNA SPQ ++ + S + N
Sbjct: 564 HNSAPQQASEQNLHP----QEPHQLDHFQPNA----AASPQPLQDSEFDSLNPSNSSNTQ 719
Query: 746 I 746
+
Sbjct: 720 L 722
>TC232895 weakly similar to PRF|1604369A.0|226743|1604369A sulfated surface
glycoprotein SSG185. {Volvox carteri;} , partial (14%)
Length = 907
Score = 39.7 bits (91), Expect = 0.006
Identities = 30/104 (28%), Positives = 43/104 (40%), Gaps = 4/104 (3%)
Frame = +3
Query: 492 PESQPAEQTTSPPHSPRSSF----FQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS 547
P S P T PP SP SS P PT +P ++P+ S S P S
Sbjct: 252 PSSPPRRNTPPPPPSPSSSTPLRPSSPPPTTSPTSR--PSRPTPSSPSPSTAPAPAPTTS 425
Query: 548 SEPIIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTA 591
+ P + + T P P T +PR RP++ T + +T+
Sbjct: 426 TTPPTPSKSKAKPTSPSPTTPTRPSPR--VRPSSFKTPSACATS 551
Score = 34.7 bits (78), Expect = 0.20
Identities = 44/164 (26%), Positives = 58/164 (34%), Gaps = 1/164 (0%)
Frame = +3
Query: 432 PTRVEPAAEVVRRTEPPARVTRSSAHSSKPALTSDDDLNLFDALPISALLQHSSNPLTPI 491
P RV A V R PP + ++ P S S L+ SS P T
Sbjct: 201 PQRVTSAT--VSRHAPPTSPSSPPRRNTPPPPPSPSS---------STPLRPSSPPPTTS 347
Query: 492 PESQPAEQTTSPPHSPRSSFFQPSPTEAPLWNLLQNQPSRSDEPTSLLTIPYDPLS-SEP 550
P S+P+ T S PSP+ AP + PT+ T P S ++P
Sbjct: 348 PTSRPSRPTPS----------SPSPSTAP-----------APAPTTSTTPPTPSKSKAKP 464
Query: 551 IIHEQPEPNQTEPQPRTSDHSAPRASARPAARTTDTDSSTAFTP 594
P + P+ R S P A A + T T S A P
Sbjct: 465 TSPSPTTPTRPSPRVRPSSFKTPSACATSSRVRTSTCLSGARVP 596
Score = 31.2 bits (69), Expect = 2.3
Identities = 26/101 (25%), Positives = 39/101 (37%), Gaps = 12/101 (11%)
Frame = +3
Query: 668 QQAHPVPDQQEPIQPDPEPEQSVSNQSSVRSPHPLVETSDPHLGTSEPNAPMINIGSPQG 727
+ A P P + P P S S+ + +R P TS TS P+ P + SP
Sbjct: 231 RHAPPTSPSSPPRRNTPPPPPSPSSSTPLRPSSPPPTTSP----TSRPSRPTPSSPSPST 398
Query: 728 A------------SEAHSSNHPASPAPNLSIIPYTHLQPTS 756
A + + S P SP+P P ++P+S
Sbjct: 399 APAPAPTTSTTPPTPSKSKAKPTSPSPTTPTRPSPRVRPSS 521
>TC226228 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement pol
polyprotein, partial (97%)
Length = 1094
Score = 39.7 bits (91), Expect = 0.006
Identities = 34/124 (27%), Positives = 58/124 (46%), Gaps = 11/124 (8%)
Frame = +2
Query: 802 QVPIMMQLLHAEGS----QSIEAAKQRFARRVALHE---LEQRNKLLEAIEEAKRKKEQA 854
Q+ I Q++ EG+ ++++A + A A+ + ++ +K ++ I E +Q
Sbjct: 458 QLRIHDQMIMLEGAKATTETVDALRTGAAAMKAMQKATNIDDVDKTMDEINEQTENMKQI 637
Query: 855 EEAARQ---AAAQAEQARLEAERLEAEAEALRCQALAPVVFTPAASASIP-DPQAAQNVP 910
++A AAA ++ LEAE E E L Q L P PAA +P Q + VP
Sbjct: 638 QDALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAAPVQVPAGQQPTRPVP 817
Query: 911 SSST 914
+ T
Sbjct: 818 AKRT 829
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,248,410
Number of Sequences: 63676
Number of extensions: 646482
Number of successful extensions: 6391
Number of sequences better than 10.0: 394
Number of HSP's better than 10.0 without gapping: 5241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6066
length of query: 952
length of database: 12,639,632
effective HSP length: 106
effective length of query: 846
effective length of database: 5,889,976
effective search space: 4982919696
effective search space used: 4982919696
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0012a.3


