

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
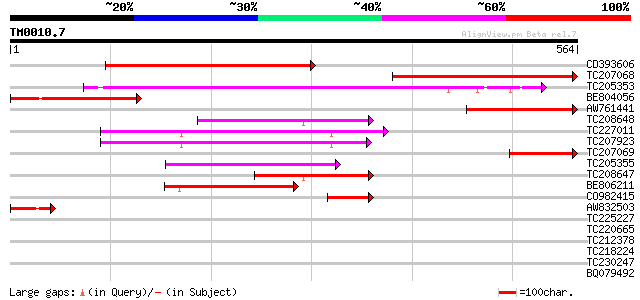
Query= TM0010.7
(564 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD393606 similar to GP|3746964|gb| signal recognition particle 5... 380 e-105
TC207068 similar to UP|O82532 (O82532) Signal recognition partic... 313 1e-85
TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 221 9e-58
BE804056 homologue to GP|3746903|gb|A signal recognition particl... 178 5e-45
AW761441 similar to GP|3746903|gb|A signal recognition particle ... 157 1e-38
TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 135 6e-32
TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 124 1e-28
TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 122 3e-28
TC207069 similar to UP|O82532 (O82532) Signal recognition partic... 115 5e-26
TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 112 6e-25
TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 100 1e-21
BE806211 similar to GP|29897491|gb Cell division protein ftsY {B... 100 3e-21
CO982415 59 6e-09
AW832503 47 2e-05
TC225227 similar to GB|AAF02830.1|6056366|AC009894 phosphoglycer... 40 0.004
TC220665 31 1.7
TC212378 similar to UP|O15324 (O15324) HPV16 E1 protein binding ... 30 3.8
TC218224 weakly similar to UP|Q84VC1 (Q84VC1) Zinc finger POZ do... 30 3.8
TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g6... 30 3.8
BQ079492 29 4.9
>CD393606 similar to GP|3746964|gb| signal recognition particle 54 kDa
subunit precursor {Arabidopsis thaliana}, partial (37%)
Length = 630
Score = 380 bits (975), Expect = e-105
Identities = 197/209 (94%), Positives = 204/209 (97%)
Frame = +3
Query: 96 GEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKI 155
GEEVL+KENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGV+PDQQLVKI
Sbjct: 3 GEEVLSKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVRPDQQLVKI 182
Query: 156 VHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGD 215
VHEELV+LMGGEVSEL FAKSG TVILLAGLQGVGKTTVCAKL+NYLKKQGKSCMLVAGD
Sbjct: 183 VHEELVQLMGGEVSELVFAKSGPTVILLAGLQGVGKTTVCAKLANYLKKQGKSCMLVAGD 362
Query: 216 IYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQID 275
+YRPAAIDQL ILGKQV VPVYTAGTDVKPSEIA+QGLEEAKKKKIDVVIVDTAGRLQID
Sbjct: 363 VYRPAAIDQLAILGKQVDVPVYTAGTDVKPSEIAKQGLEEAKKKKIDVVIVDTAGRLQID 542
Query: 276 KAMMDELKEVKKSLNPTEVLLVVDAMTGQ 304
K MMDELKEVKK+LNPTEVLLVVDAMTGQ
Sbjct: 543 KTMMDELKEVKKALNPTEVLLVVDAMTGQ 629
>TC207068 similar to UP|O82532 (O82532) Signal recognition particle 54 kDa
subunit (Fragment), partial (33%)
Length = 930
Score = 313 bits (803), Expect = 1e-85
Identities = 162/185 (87%), Positives = 175/185 (94%), Gaps = 1/185 (0%)
Frame = +1
Query: 381 VEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTPAQ 440
VEKAQ+V +QEDAE+LQ+KIMSAKFDFNDFLKQTR VA+MGSVSRV+GMIPGM KVTPAQ
Sbjct: 1 VEKAQQVXRQEDAEELQKKIMSAKFDFNDFLKQTRTVAKMGSVSRVIGMIPGMGKVTPAQ 180
Query: 441 IREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQM 500
IR+AEKNL NMEAMIEAMTPEEREKPELLAESPVRRKRVAQ+SGKTEQQVSQ+VAQLFQM
Sbjct: 181 IRDAEKNLLNMEAMIEAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQVSQLVAQLFQM 360
Query: 501 RVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRSLFADSA-ARPTPR 559
RVRMKNLMGVM+ GS+PTLSNLEEALK EEKAPPGTARRRKRSE RS+F DS ARP+PR
Sbjct: 361 RVRMKNLMGVMESGSLPTLSNLEEALKAEEKAPPGTARRRKRSESRSVFGDSTPARPSPR 540
Query: 560 GFGSK 564
GFGSK
Sbjct: 541 GFGSK 555
>TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), complete
Length = 1889
Score = 221 bits (562), Expect = 9e-58
Identities = 139/468 (29%), Positives = 246/468 (51%), Gaps = 7/468 (1%)
Frame = +3
Query: 74 VVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSV 133
+V AE+ G ++ L+ ++ V+ ++ + + + I RALL++DV +VR ++
Sbjct: 66 MVLAELGGSISRALQ----QMSNATVIDEKVLNDCLNVITRALLQSDVQFKLVRDLQTNI 233
Query: 134 SDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTT 193
+ + G + + + V EL K++ K T+V++ GLQG GKTT
Sbjct: 234 KNIVNLDDLAAGHNKRRIIQQAVFNELCKILDPGKPSFTPKKGKTSVVMFVGLQGSGKTT 413
Query: 194 VCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGL 253
C K + Y +K+G LV D +R A DQL + +P Y + + P +IA +G+
Sbjct: 414 TCTKYAYYHQKKGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGV 593
Query: 254 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTF 313
E KK+ D++IVDT+GR + + A+ +E+++V ++ P V+ V+D+ GQ A F
Sbjct: 594 ERFKKENCDLIIVDTSGRHKQEAALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAF 773
Query: 314 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILG 373
+ + I+TK+DG ++GG ALS + P+ +G GE MD+ E F R+LG
Sbjct: 774 KQSVAVGAVIVTKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 953
Query: 374 MGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM 433
MGD F++K EV+ + +L +K+ F +Q + + +MG +S+V M+PG
Sbjct: 954 MGDWSGFMDKIHEVVPMDQQPELLQKLSEGNFTLRIMYEQFQNILKMGPISQVFSMLPGF 1133
Query: 434 SK--VTPAQIREAEKNLKNMEAMIEAMTPEERE--KPELLAESPVRRKRVAQESGKTEQQ 489
S + + +E++ +K M+++MT EE + P+L+ ES R R+A+ SG+ ++
Sbjct: 1134SAELMPKGREKESQAKIKRYMTMMDSMTNEELDSSNPKLMNES--RMMRIARGSGRPVRE 1307
Query: 490 VSQVVAQ---LFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPP 534
V +++ + L ++ +MK L + + G M LS A + PP
Sbjct: 1308VMEMLEEYKRLAKIWSKMKGLK-IPKKGEMSALSRNMNAQHMSKVLPP 1448
>BE804056 homologue to GP|3746903|gb|A signal recognition particle 54 kDa
subunit precursor {Pisum sativum}, partial (11%)
Length = 391
Score = 178 bits (452), Expect = 5e-45
Identities = 96/132 (72%), Positives = 106/132 (79%), Gaps = 1/132 (0%)
Frame = +1
Query: 1 MEAVLASRHFSTPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASISSRNSFTKEIWGWVQA 60
+EAV SRHFST S S N+T+ SSS S+KLSSPWTNAS++S N F KE+WGWV +
Sbjct: 1 VEAVAGSRHFSTLPLSFSQNKTLRSSSAK--SIKLSSPWTNASLTSPNCFAKEVWGWVNS 174
Query: 61 KSVAVRRRRDMPGVV-RAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEA 119
S V RRD GVV RAEMFGQLT+GLE AWNKLKGEEVL+KENIVEPMRDI RALLEA
Sbjct: 175 NSKGVAVRRDTRGVVVRAEMFGQLTSGLETAWNKLKGEEVLSKENIVEPMRDIXRALLEA 354
Query: 120 DVSLPVVRRFVQ 131
DVSLPVVRRFVQ
Sbjct: 355 DVSLPVVRRFVQ 390
>AW761441 similar to GP|3746903|gb|A signal recognition particle 54 kDa
subunit precursor {Pisum sativum}, partial (16%)
Length = 364
Score = 157 bits (397), Expect = 1e-38
Identities = 79/111 (71%), Positives = 95/111 (85%), Gaps = 1/111 (0%)
Frame = +1
Query: 455 IEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGG 514
IEAMTPEEREKPELLAESPVRRKRVAQ+S KT+ V Q+VAQLFQMRVRMKNLMGVM+ G
Sbjct: 1 IEAMTPEEREKPELLAESPVRRKRVAQDSRKTDHHVXQLVAQLFQMRVRMKNLMGVMESG 180
Query: 515 SMPTLSNLEEALKPEEKAPPGTARRRKRSEQRSLFADSA-ARPTPRGFGSK 564
S+PTLSNLEEALK EE APP TA+++K+ + +++F DS A+P+P F +K
Sbjct: 181 SLPTLSNLEEALKAEENAPPDTAKKKKKCKSKNVFRDSTPAKPSPXSFRNK 333
>TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (53%)
Length = 845
Score = 135 bits (339), Expect = 6e-32
Identities = 75/182 (41%), Positives = 108/182 (59%), Gaps = 7/182 (3%)
Frame = +1
Query: 188 GVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTD-VKPS 246
G GKTT KL+ LK +G ++ AGD +R AA DQL I + G + A ++ K S
Sbjct: 7 GGGKTTSLGKLAYRLKNEGAKILMAAGDTFRAAASDQLEIWAGRTGCEIVVAESEKAKAS 186
Query: 247 EIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLN------PTEVLLVVDA 300
+ Q +++ K+ D+V+ DT+GRL + ++M+EL KKS+ P E+LLV+D
Sbjct: 187 SVLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLVLDG 366
Query: 301 MTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLE 360
TG FN +G+TG ILTKLDG +RGG +SV + G P+K VG GE ++DL+
Sbjct: 367 TTGLNMLPQAREFNDVVGVTGLILTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVEDLQ 546
Query: 361 PF 362
PF
Sbjct: 547 PF 552
>TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(55%)
Length = 1496
Score = 124 bits (311), Expect = 1e-28
Identities = 82/304 (26%), Positives = 156/304 (50%), Gaps = 18/304 (5%)
Frame = +3
Query: 91 WNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQ 150
+ + G+ L K ++ ++ ++ L+ +V+ + + +SV+ G + +
Sbjct: 318 FQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISS 497
Query: 151 QLVKIVHEELVKLMGGEVS-------ELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLK 203
+ + E LV+++ S A + V++ G+ GVGK+T AK++ +L
Sbjct: 498 TVHTAMEEALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLL 677
Query: 204 KQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDV 263
+ S M+ A D +R A++QL +++ +P++ G + P+ +A++ ++EA + DV
Sbjct: 678 QHNVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAIVAKEAIQEAARNGSDV 857
Query: 264 VIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG----- 318
V+VDTAGR+Q ++ +M L ++ NP +L V +A+ G +A ++ FN ++
Sbjct: 858 VLVDTAGRMQDNEPLMRALSKLVYLNNPDLILFVGEALVGNDAVDQLSKFNQKLADLSTS 1037
Query: 319 -----ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRIL 372
I G +LTK D D + GAALS+ VSG P+ VG G+ DL+ +A +L
Sbjct: 1038PTPRLIDGILLTKFDTIDDKVGAALSMVYVSGAPVMFVGCGQSYTDLKKLNVKSIAKTLL 1217
Query: 373 GMGD 376
G+
Sbjct: 1218K*GN 1229
>TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(60%)
Length = 1569
Score = 122 bits (307), Expect = 3e-28
Identities = 78/288 (27%), Positives = 150/288 (52%), Gaps = 18/288 (6%)
Frame = +2
Query: 91 WNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQ 150
+ + G+ L K ++ ++ ++ L+ +V+ + + +SV+ G + +
Sbjct: 455 FQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISS 634
Query: 151 QLVKIVHEELVKLMGGEVS-------ELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLK 203
+ + E L++++ S A + V++ G+ GVGK+T AK++ +L
Sbjct: 635 TVHAAMEEALIRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLL 814
Query: 204 KQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDV 263
+ S M+ A D +R A++QL +++ +P++ G + P+ +A++ ++EA + DV
Sbjct: 815 QHKVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDV 994
Query: 264 VIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG----- 318
V+VDTAGR+Q ++ +M L ++ NP VL V +A+ G +A ++ FN ++
Sbjct: 995 VLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLATS 1174
Query: 319 -----ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMDDLE 360
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 1175PNPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 1318
>TC207069 similar to UP|O82532 (O82532) Signal recognition particle 54 kDa
subunit (Fragment), partial (8%)
Length = 578
Score = 115 bits (288), Expect = 5e-26
Identities = 58/68 (85%), Positives = 62/68 (90%), Gaps = 1/68 (1%)
Frame = +3
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRSLFADSA-ARP 556
FQMRVRMKNLMGVM+ GS+PTLSNLEEALK EEKAPPGTARRRKRSE RS+F DS ARP
Sbjct: 3 FQMRVRMKNLMGVMESGSLPTLSNLEEALKAEEKAPPGTARRRKRSESRSVFGDSTPARP 182
Query: 557 TPRGFGSK 564
+PRGFGSK
Sbjct: 183 SPRGFGSK 206
>TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), partial (37%)
Length = 554
Score = 112 bits (279), Expect = 6e-25
Identities = 59/174 (33%), Positives = 96/174 (54%)
Frame = +3
Query: 156 VHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGD 215
V EL K++ K T+V++ GLQG GKTT C K + Y +K+G LV D
Sbjct: 33 VFNELCKILDPGKPSFTPKKGKTSVVMFVGLQGSGKTTTCTKYAFYHQKKGWKPALVCAD 212
Query: 216 IYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQID 275
+R A DQL + +P Y + + P +IA +G+E K++ D++IVDT+GR + +
Sbjct: 213 TFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGVERFKQENCDLIIVDTSGRHKQE 392
Query: 276 KAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDG 329
A+ +E+++V ++ P V+ V+D+ GQ A F + + I+TK+DG
Sbjct: 393 AALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAFKQSVAVGAVIVTKMDG 554
>TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (37%)
Length = 817
Score = 100 bits (250), Expect = 1e-21
Identities = 52/125 (41%), Positives = 77/125 (61%), Gaps = 6/125 (4%)
Frame = +1
Query: 244 KPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLN------PTEVLLV 297
K S + Q +++ K+ D+V+ DT+GRL + ++M+EL KKS+ P E+LLV
Sbjct: 1 KASSVLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLV 180
Query: 298 VDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMD 357
+D TG FN +G+TG +LTKLDG +RGG +SV + G P+K VG GE ++
Sbjct: 181 LDGTTGLNMLPQAREFNDVVGVTGLVLTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVE 360
Query: 358 DLEPF 362
DL+PF
Sbjct: 361 DLQPF 375
>BE806211 similar to GP|29897491|gb Cell division protein ftsY {Bacillus
cereus ATCC 14579}, partial (43%)
Length = 436
Score = 99.8 bits (247), Expect = 3e-21
Identities = 52/136 (38%), Positives = 84/136 (61%), Gaps = 3/136 (2%)
Frame = -1
Query: 155 IVHEELVKLMGGE---VSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCML 211
++ ++L+K+ G +E+ K G+T +L G+ GVGKTT K+++ +GKS +L
Sbjct: 412 VISKKLIKIYTGTSDFTNEINEPKDGSTDVLFVGVNGVGKTTTIGKMAHKF*SEGKSVLL 233
Query: 212 VAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGR 271
AGD +R AI+QL + G +VGV V G+ P+ + ++ AK +K+DV++ DTAGR
Sbjct: 232 AAGDTFRDGAIEQL*VSGDRVGVEVIKQGSGSDPAAVMYDAVQAAKARKVDVLLCDTAGR 53
Query: 272 LQIDKAMMDELKEVKK 287
LQ +M EL +VK+
Sbjct: 52 LQNKVNLMKELDKVKR 5
>CO982415
Length = 806
Score = 58.9 bits (141), Expect = 6e-09
Identities = 26/46 (56%), Positives = 35/46 (75%)
Frame = -2
Query: 317 IGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPF 362
+G+TG ILTKLDG +RGG +SV + G P+K VG GE ++DL+PF
Sbjct: 523 VGVTGLILTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVEDLQPF 386
>AW832503
Length = 303
Score = 47.4 bits (111), Expect = 2e-05
Identities = 27/45 (60%), Positives = 33/45 (73%)
Frame = +3
Query: 1 MEAVLASRHFSTPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASIS 45
MEAV S HFST + S SHN+T+ SS S S++LSSP TNAS+S
Sbjct: 48 MEAVTGSHHFSTLSLSFSHNKTLRSS--SAKSIRLSSPSTNASLS 176
>TC225227 similar to GB|AAF02830.1|6056366|AC009894 phosphoglycerate kinase
{Arabidopsis thaliana;} , partial (35%)
Length = 612
Score = 39.7 bits (91), Expect = 0.004
Identities = 44/169 (26%), Positives = 82/169 (48%), Gaps = 7/169 (4%)
Frame = +1
Query: 9 HFSTPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASISSRNSFTKE---IWGWVQAKSVAV 65
H +TP SSS ++S SP+ ++LSS + ++ R F + V A+ AV
Sbjct: 133 HSTTPASSSS------ATSASPSRVQLSSSLRSRPLARRLGFAAADPLLAANVAARVAAV 294
Query: 66 RRRRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADV--SL 123
R + GVV M + L AA LKG++V + ++ P+ D + + + ++
Sbjct: 295 RGGKGARGVV--SMAKKSVGELSAA--DLKGKKVFVRADLNVPLDDNQNITDDTRIRAAI 462
Query: 124 PVVRRFVQSVSDQAVG--VGVIRGVKPDQQLVKIVHEELVKLMGGEVSE 170
P ++ +Q+ + + +G +GV P L +V L +L+G +V++
Sbjct: 463 PTIKHLIQNGAKVILSSHLGRPKGVTPKYSLAPLV-PRLSELIGXQVAK 606
>TC220665
Length = 699
Score = 30.8 bits (68), Expect = 1.7
Identities = 20/48 (41%), Positives = 26/48 (53%), Gaps = 8/48 (16%)
Frame = -3
Query: 10 FSTPTHSS--------SHNRTVCSSSPSPNSLKLSSPWTNASISSRNS 49
FS PTH S SH T+ +SSPSPN L+ ++ T +S NS
Sbjct: 310 FSKPTHLSKQPLWKTCSHLITLITSSPSPNLLRHTAQSTCSSEPEENS 167
>TC212378 similar to UP|O15324 (O15324) HPV16 E1 protein binding protein
(Thyroid hormone receptor interactor 13) (TRIP13
protein), partial (36%)
Length = 1106
Score = 29.6 bits (65), Expect = 3.8
Identities = 29/96 (30%), Positives = 46/96 (47%), Gaps = 5/96 (5%)
Frame = +1
Query: 112 IRRALLEADV--SLP-VVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEV 168
++RAL E V +LP VV+ + S + G +KP ++ V++ L V
Sbjct: 13 LQRALAETPVVNTLPRVVKHSLLETSP*SSGAQTPLSLKPRREAVELAS*RLSPSY*PRV 192
Query: 169 --SELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYL 202
A +S ++ILL G G GKT++C L+ L
Sbjct: 193 VCPWKARLRSSYSIILLQGPPGTGKTSLCKALAQKL 300
>TC218224 weakly similar to UP|Q84VC1 (Q84VC1) Zinc finger POZ domain protein
(Fragment), partial (60%)
Length = 766
Score = 29.6 bits (65), Expect = 3.8
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 3/34 (8%)
Frame = -3
Query: 8 RHFSTPTHSSSHNRTVCSSSPSPN---SLKLSSP 38
RHF +P +SSSH TV SSS SL+ S P
Sbjct: 278 RHFPSPLNSSSHPATVFSSSDCSKGQFSLRYSKP 177
>TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61870
{Arabidopsis thaliana;} , partial (56%)
Length = 1291
Score = 29.6 bits (65), Expect = 3.8
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = +1
Query: 11 STPTHSSSHNRTVCSSSPSPNSLKLSSP 38
S+PT SSS R +CS +PS S K S P
Sbjct: 325 SSPTPSSSTARPICSITPSAPSPKTSPP 408
>BQ079492
Length = 426
Score = 29.3 bits (64), Expect = 4.9
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = +3
Query: 11 STPTHSSSHNRTVCSSSPSPNSLKLSSPWTNASISS 46
+TP+ SSS + + S++PSP+S SSP S +S
Sbjct: 279 TTPSSSSSSSSSPSSATPSPSSSSSSSPPPGTSSTS 386
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,204,968
Number of Sequences: 63676
Number of extensions: 206813
Number of successful extensions: 1735
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1712
length of query: 564
length of database: 12,639,632
effective HSP length: 102
effective length of query: 462
effective length of database: 6,144,680
effective search space: 2838842160
effective search space used: 2838842160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0010.7


