

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.21
(161 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
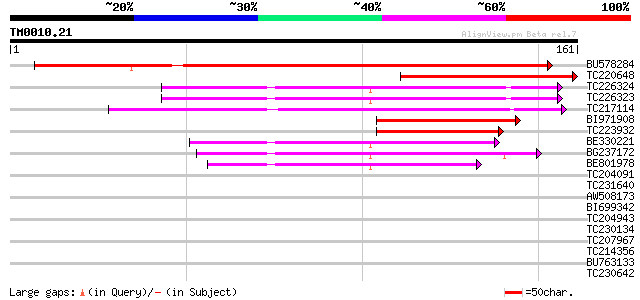
Score E
Sequences producing significant alignments: (bits) Value
BU578284 218 8e-58
TC220648 94 3e-20
TC226324 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (71%) 73 5e-14
TC226323 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (72%) 73 7e-14
TC217114 similar to UP|Q944M8 (Q944M8) Iron-sulfur cluster assem... 65 2e-11
BI971908 64 3e-11
TC223932 63 7e-11
BE330221 similar to PIR|F86238|F862 protein T10O24.11 [imported]... 62 2e-10
BG237172 weakly similar to PIR|F86238|F86 protein T10O24.11 [imp... 50 6e-07
BE801978 weakly similar to PIR|F86238|F862 protein T10O24.11 [im... 42 1e-04
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 31 0.22
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 30 0.50
AW508173 30 0.50
BI699342 30 0.65
TC204943 similar to UP|Q8L985 (Q8L985) Thylakoid lumenal 16.5 kD... 30 0.65
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 29 0.85
TC207967 similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,... 29 1.1
TC214356 similar to UP|P93704 (P93704) HMG-1, partial (44%) 28 1.4
BU763133 similar to GP|14253163|em VMP4 protein {Volvox carteri ... 28 1.4
TC230642 weakly similar to UP|O81444 (O81444) Kinase associated ... 28 1.4
>BU578284
Length = 443
Score = 218 bits (556), Expect = 8e-58
Identities = 116/150 (77%), Positives = 125/150 (83%), Gaps = 3/150 (2%)
Frame = +3
Query: 8 RRLAPSLAARFRQNQKLTSFSSSSSS---SVLHDATTSSSSSSHVEPVHMTENCLRRMKE 64
R LA LAARFR N L SSSSS+ S LH AT S SSS EPVH+T+NC+RRMKE
Sbjct: 3 RPLALHLAARFRHNHNLVRLSSSSSAASASTLHHATPSPSSS---EPVHITQNCVRRMKE 173
Query: 65 LEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINSDDRVFEREGIKLVVDNISYDFVK 124
LE SES+T LRLSVETGGCSGFQY F+LD+RINSDD+VFEREG+KLVVDNISYDFVK
Sbjct: 174 LEGSESSTDGNLLRLSVETGGCSGFQYVFNLDNRINSDDKVFEREGVKLVVDNISYDFVK 353
Query: 125 GATVDYVEELIRSAFVVTENPSAVGGCSCK 154
GATVDYVEELIRSAFVVTENPSAVGGCSCK
Sbjct: 354 GATVDYVEELIRSAFVVTENPSAVGGCSCK 443
>TC220648
Length = 468
Score = 94.0 bits (232), Expect = 3e-20
Identities = 46/50 (92%), Positives = 46/50 (92%)
Frame = +3
Query: 112 KLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAVGGCSCKSSFMVKQ 161
KLVVDNISYDFVKGATVDYVEELIRS TENPSAVGGCSCKSSFMVKQ
Sbjct: 3 KLVVDNISYDFVKGATVDYVEELIRSXXXXTENPSAVGGCSCKSSFMVKQ 152
>TC226324 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (71%)
Length = 844
Score = 73.2 bits (178), Expect = 5e-14
Identities = 40/116 (34%), Positives = 62/116 (52%), Gaps = 2/116 (1%)
Frame = +3
Query: 44 SSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS-- 101
+S S + +T+N L+ + ++ + + LR+ V+ GGCSG Y D +DR+N
Sbjct: 216 ASGSLAPAISVTDNVLKHLNKMRSERNQDL--CLRIGVKQGGCSGMSYTMDFEDRVNKRP 389
Query: 102 DDRVFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAVGGCSCKSSF 157
DD + E EG ++V D S F+ G +DY + LI F +NP+A C C SF
Sbjct: 390 DDSIIEYEGFEIVCDPKSLLFIFGMQLDYSDALIGGGFSF-KNPNATQTCGCGKSF 554
>TC226323 similar to UP|Q6NM99 (Q6NM99) At1g10500, partial (72%)
Length = 810
Score = 72.8 bits (177), Expect = 7e-14
Identities = 41/116 (35%), Positives = 61/116 (52%), Gaps = 2/116 (1%)
Frame = +3
Query: 44 SSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS-- 101
+S S V +T+N L+ + ++ + + LR+ V+ GGCSG Y D +DR N
Sbjct: 255 ASGSLAPAVSLTDNALKHLNKMRSERNQDL--CLRIGVKQGGCSGMSYTMDFEDRANKRP 428
Query: 102 DDRVFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAVGGCSCKSSF 157
DD + E EG ++V D S F+ G +DY + LI F +NP+A C C SF
Sbjct: 429 DDSIIEYEGFEIVCDPKSLLFIFGMQLDYSDALIGGGFSF-KNPNATQTCGCGKSF 593
>TC217114 similar to UP|Q944M8 (Q944M8) Iron-sulfur cluster assembly protein
IscA, partial (64%)
Length = 925
Score = 64.7 bits (156), Expect = 2e-11
Identities = 36/130 (27%), Positives = 67/130 (50%)
Frame = +2
Query: 29 SSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETGGCSG 88
SS +S + A ++++ + V +T+ R+++L L+L V+ GC+G
Sbjct: 107 SSMASKAMSVAAEKIATAARRQAVSLTDAAASRIRQLLQQRQRPF---LKLGVKARGCNG 277
Query: 89 FQYAFDLDDRINSDDRVFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAV 148
Y + D D + E +G+K+++D + V G +D+V++ +RS F+ NP++
Sbjct: 278 LSYTLNYADEKGKFDELVEDKGVKILIDPKALMHVIGTKMDFVDDKLRSEFIFV-NPNSK 454
Query: 149 GGCSCKSSFM 158
G C C SFM
Sbjct: 455 GQCGCGESFM 484
>BI971908
Length = 420
Score = 63.9 bits (154), Expect = 3e-11
Identities = 31/41 (75%), Positives = 35/41 (84%)
Frame = +2
Query: 105 VFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENP 145
VFER+GIKLVVDNISYDFVKGATVDYVEEL +F+ + P
Sbjct: 167 VFERDGIKLVVDNISYDFVKGATVDYVEELSSHSFLHSGTP 289
Score = 33.1 bits (74), Expect = 0.059
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +1
Query: 39 ATTSSSSSSHVEPVHMTENCLR 60
AT+SS S +EP+HMT+NC+R
Sbjct: 100 ATSSSDSDPDLEPLHMTKNCVR 165
>TC223932
Length = 530
Score = 62.8 bits (151), Expect = 7e-11
Identities = 30/36 (83%), Positives = 33/36 (91%)
Frame = +2
Query: 105 VFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFV 140
VFER+GIKLVVDNISYDFVKGATVDYVEEL +F+
Sbjct: 182 VFERDGIKLVVDNISYDFVKGATVDYVEELSSHSFL 289
Score = 33.1 bits (74), Expect = 0.059
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +1
Query: 39 ATTSSSSSSHVEPVHMTENCLR 60
AT+SS S +EP+HMT+NC+R
Sbjct: 115 ATSSSDSDPDLEPLHMTKNCVR 180
>BE330221 similar to PIR|F86238|F862 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (79%)
Length = 295
Score = 61.6 bits (148), Expect = 2e-10
Identities = 31/90 (34%), Positives = 52/90 (57%), Gaps = 2/90 (2%)
Frame = +2
Query: 52 VHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS--DDRVFERE 109
V +T+N L+ + ++ + + LR+ V+ GGCSG Y D++DR N DD + E E
Sbjct: 32 VALTDNALKHLNKMRSERNQDL--CLRICVKHGGCSGMSYTMDVEDRANKRPDDAIMENE 205
Query: 110 GIKLVVDNISYDFVKGATVDYVEELIRSAF 139
G+++V D S ++ G +DY + LI +F
Sbjct: 206 GLEIVCDP*SLLYIYGVQLDYSDALIGGSF 295
>BG237172 weakly similar to PIR|F86238|F86 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (92%)
Length = 340
Score = 49.7 bits (117), Expect = 6e-07
Identities = 30/101 (29%), Positives = 48/101 (46%), Gaps = 3/101 (2%)
Frame = +2
Query: 54 MTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS--DDRVFEREGI 111
+T+N L+ + ++ + + +RL V GGCS Y D +DR N DD + E EG
Sbjct: 20 LTDNALKHLNKMRSERNQDL--CVRLGVRHGGCSAMSYTMDSEDRANKTPDDSIIEYEGC 193
Query: 112 KLVVDNISYDFVKGATVDYVEELIRSAF-VVTENPSAVGGC 151
++ D + G +DY + LI + T N + GC
Sbjct: 194 EIDSDPKILLLILGMQLDYSDALIVGGISIKTPNATQTRGC 316
>BE801978 weakly similar to PIR|F86238|F862 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (70%)
Length = 250
Score = 42.0 bits (97), Expect = 1e-04
Identities = 23/80 (28%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Frame = +2
Query: 57 NCLRRMKELEASESATAPKTLRLSVETGGCSGFQYAFDLDDRINS--DDRVFEREGIKLV 114
N L+ + + + + +R+ V+ GGCSG Y D +DR+N +D + E E + V
Sbjct: 2 NALKHLNNMRSERNQDL--CVRIGVKQGGCSGMSYTMDFEDRVNKGPNDAIIEYECKEKV 175
Query: 115 VDNISYDFVKGATVDYVEEL 134
D S F+ +D+ + L
Sbjct: 176 CDPNSLLFILDMQIDHSDAL 235
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 31.2 bits (69), Expect = 0.22
Identities = 24/65 (36%), Positives = 32/65 (48%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETG 84
TSFSSSSSSS +++SSSSSS +S S+++ + S G
Sbjct: 252 TSFSSSSSSSSSSSSSSSSSSSS--------------SSSTSSSTSSSSTSSSTSSYS*G 115
Query: 85 GCSGF 89
G SGF
Sbjct: 114 GFSGF 100
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 30.0 bits (66), Expect = 0.50
Identities = 19/39 (48%), Positives = 26/39 (65%), Gaps = 1/39 (2%)
Frame = -3
Query: 10 LAPSLAAR-FRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
L+ S+ R F + +SFSSSSSSS +++SSSSSS
Sbjct: 164 LSSSMITRTFPSSSSSSSFSSSSSSSSSSSSSSSSSSSS 48
Score = 28.5 bits (62), Expect = 1.4
Identities = 18/45 (40%), Positives = 27/45 (60%)
Frame = -3
Query: 3 TSSLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
+SS+ R PS ++ + +S SSSSSSS +++S SSSS
Sbjct: 161 SSSMITRTFPSSSSSSSFSSSSSSSSSSSSSSSSSSSSSSFSSSS 27
>AW508173
Length = 226
Score = 30.0 bits (66), Expect = 0.50
Identities = 17/25 (68%), Positives = 19/25 (76%)
Frame = -1
Query: 23 KLTSFSSSSSSSVLHDATTSSSSSS 47
KLTS S SSSSS L ++ SSSSSS
Sbjct: 142 KLTSESQSSSSSSLSSSSNSSSSSS 68
>BI699342
Length = 325
Score = 29.6 bits (65), Expect = 0.65
Identities = 16/25 (64%), Positives = 20/25 (80%)
Frame = +3
Query: 23 KLTSFSSSSSSSVLHDATTSSSSSS 47
KL +FSSSSSSS +++SSSSSS
Sbjct: 177 KLIAFSSSSSSSSSSSSSSSSSSSS 251
Score = 28.5 bits (62), Expect = 1.4
Identities = 16/30 (53%), Positives = 21/30 (69%)
Frame = +3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHM 54
+S SSSSSSS +++SSSSSS E H+
Sbjct: 198 SSSSSSSSSSSSSSSSSSSSSSSAFEVYHL 287
>TC204943 similar to UP|Q8L985 (Q8L985) Thylakoid lumenal 16.5 kDa protein,
chloroplast, partial (68%)
Length = 1055
Score = 29.6 bits (65), Expect = 0.65
Identities = 21/96 (21%), Positives = 47/96 (48%), Gaps = 3/96 (3%)
Frame = +2
Query: 29 SSSSSSVLHDATTSSSSSSHVEPVHMTEN-CLRRMKELEASESATAPKTLRLSVETGGCS 87
S+S+ +L ++ SSSSSS V+P+H+ N ++ + E + P + + + G
Sbjct: 38 STSNPILLPSSSPSSSSSSFVKPIHVCSNPIIKSTSKRELTLCKATPHSPIVVTKRGLSI 217
Query: 88 GF--QYAFDLDDRINSDDRVFEREGIKLVVDNISYD 121
F + L + N++ + E + + +++ + D
Sbjct: 218 SFITSFVLSLAGKKNANAAILEADDDEELLEKVKRD 325
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 29.3 bits (64), Expect = 0.85
Identities = 19/50 (38%), Positives = 29/50 (58%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAP 74
+S SS SSSS+ +++SSSSSS + P + +SESA++P
Sbjct: 495 SSSSSPSSSSLSSSSSSSSSSSSSLSPPPLASG--------SSSESASSP 370
>TC207967 similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,
chloroplast precursor (TRX-M3), partial (56%)
Length = 528
Score = 28.9 bits (63), Expect = 1.1
Identities = 18/37 (48%), Positives = 21/37 (56%), Gaps = 9/37 (24%)
Frame = +2
Query: 24 LTSFSSSSSSSVLHDAT---------TSSSSSSHVEP 51
+ SFS +SSS LH AT +SSSSSSH P
Sbjct: 29 MASFSLTSSSPTLHKATIIDPSSSSSSSSSSSSHAFP 139
>TC214356 similar to UP|P93704 (P93704) HMG-1, partial (44%)
Length = 640
Score = 28.5 bits (62), Expect = 1.4
Identities = 16/32 (50%), Positives = 19/32 (59%)
Frame = -1
Query: 20 QNQKLTSFSSSSSSSVLHDATTSSSSSSHVEP 51
QN +L +SSSSSS+ D S SSSS P
Sbjct: 322 QNNQLDQSTSSSSSSLTSDFDLSDSSSSEFFP 227
>BU763133 similar to GP|14253163|em VMP4 protein {Volvox carteri f.
nagariensis}, partial (1%)
Length = 452
Score = 28.5 bits (62), Expect = 1.4
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = +3
Query: 20 QNQKLTSFSSSSSSSVLHDATTSSSSSS 47
+N+K+ S + S+SS+ TTSSSSSS
Sbjct: 288 ENEKIGSHAKSTSSTPAWQTTTSSSSSS 371
>TC230642 weakly similar to UP|O81444 (O81444) Kinase associated protein
phosphatase, partial (28%)
Length = 609
Score = 28.5 bits (62), Expect = 1.4
Identities = 16/34 (47%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Frame = -1
Query: 4 SSLFRRLAPSLAARFRQNQKL-TSFSSSSSSSVL 36
S FR L+PS+ ARF +++ +SFS S SS+L
Sbjct: 390 SCCFRNLSPSILARFIPYRRVSSSFSGSPVSSIL 289
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.126 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,805,279
Number of Sequences: 63676
Number of extensions: 105901
Number of successful extensions: 2076
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 1241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1648
length of query: 161
length of database: 12,639,632
effective HSP length: 90
effective length of query: 71
effective length of database: 6,908,792
effective search space: 490524232
effective search space used: 490524232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0010.21


