

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.2
(370 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
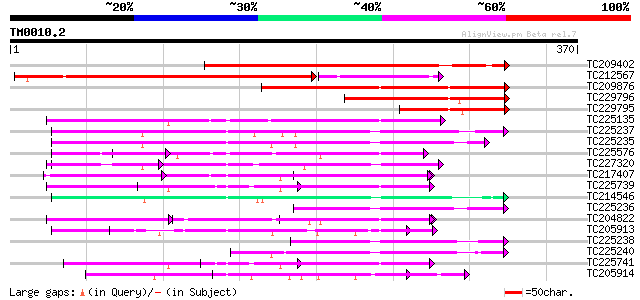
Score E
Sequences producing significant alignments: (bits) Value
TC209402 322 1e-88
TC212567 weakly similar to UP|PAB3_HUMAN (Q9H361) Polyadenylate-... 280 7e-76
TC209876 similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A10_14, ... 179 2e-45
TC229796 163 1e-40
TC229795 weakly similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A... 96 3e-20
TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, ... 87 9e-18
TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 83 2e-16
TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {A... 79 4e-15
TC225576 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, ... 74 1e-13
TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding prot... 72 3e-13
TC217407 similar to UP|P82277 (P82277) Plastid-specific 30S ribo... 66 3e-11
TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 65 5e-11
TC214546 similar to PIR|T01932|T01932 RNA binding protein homolo... 64 8e-11
TC225236 similar to UP|Q84LL7 (Q84LL7) Salt tolerance protein 6,... 64 1e-10
TC204822 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, part... 63 2e-10
TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protei... 62 3e-10
TC225238 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 62 3e-10
TC225240 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 62 4e-10
TC225741 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 60 1e-09
TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, ... 60 2e-09
>TC209402
Length = 1163
Score = 322 bits (826), Expect = 1e-88
Identities = 162/199 (81%), Positives = 177/199 (88%)
Frame = +1
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
++GPGVI +ELLKD Q+S RNRG+AFIEYYNHACAEYSRQKMSNSNFKL +NAPTVSWA+
Sbjct: 1 EIGPGVICVELLKDPQNSSRNRGYAFIEYYNHACAEYSRQKMSNSNFKLGSNAPTVSWAD 180
Query: 188 PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVH 247
PRNSESSA+S VK+VYVKNLPENITQD LK+LFEHHGKITKVVLP AKSGQEKSRFGFVH
Sbjct: 181 PRNSESSAISLVKSVYVKNLPENITQDRLKELFEHHGKITKVVLPSAKSGQEKSRFGFVH 360
Query: 248 FSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGM 307
F++RSSAMKALKNTEKYEIDG+ LECSLAKP+A NSQKP +LPTYP LGYGM
Sbjct: 361 FAERSSAMKALKNTEKYEIDGQLLECSLAKPQA--------NSQKPALLPTYPPHLGYGM 516
Query: 308 VGGAYGGIGAGYGAAGFAQ 326
VGGA IGAGYGAAGFAQ
Sbjct: 517 VGGA---IGAGYGAAGFAQ 564
Score = 34.3 bits (77), Expect = 0.092
Identities = 30/156 (19%), Positives = 70/156 (44%), Gaps = 14/156 (8%)
Frame = +1
Query: 55 VRIMKAKESGER-KGYAFVAFKTKELASQAIEELNNSEFK------------GKKIKCSS 101
V ++K ++ R +GYAF+ + A + ++++NS FK + + S+
Sbjct: 25 VELLKDPQNSSRNRGYAFIEYYNHACAEYSRQKMSNSNFKLGSNAPTVSWADPRNSESSA 204
Query: 102 SQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHAC 161
+++ N+P+ T + +K++ G I+ +L +S F F+ + +
Sbjct: 205 ISLVKSVYVKNLPENITQDRLKELFEH--HGKITKVVLPSAKSGQEKSRFGFVHFAERSS 378
Query: 162 AEYSRQKMSNSNFKLENNAPTVSWAEPR-NSESSAV 196
A + + ++++ S A+P+ NS+ A+
Sbjct: 379 A--MKALKNTEKYEIDGQLLECSLAKPQANSQKPAL 480
>TC212567 weakly similar to UP|PAB3_HUMAN (Q9H361) Polyadenylate-binding
protein 3 (Poly(A)-binding protein 3) (PABP 3)
(Testis-specific poly(A)-binding protein), partial (5%)
Length = 615
Score = 280 bits (716), Expect = 7e-76
Identities = 134/203 (66%), Positives = 165/203 (81%), Gaps = 6/203 (2%)
Frame = +3
Query: 4 NHTNDEV------EKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRI 57
NH++DE EK+KHAELL+LPPH SEVYIGGI + S+EDL+ C+ +GEV+EVRI
Sbjct: 9 NHSSDEAKVEDEDEKRKHAELLSLPPHGSEVYIGGIPH-ASDEDLKSLCERIGEVAEVRI 185
Query: 58 MKAKESGERKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKW 117
MK K E KG+ FV F + ELAS+AIEELNN+EF GKKIKCS SQAKHRLFIGN+P+ W
Sbjct: 186 MKGKXFFENKGFGFVTFTSVELASKAIEELNNTEFMGKKIKCSKSQAKHRLFIGNVPRSW 365
Query: 118 TVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLE 177
VED+KK+V ++GPGV +EL+KD++++ NRGFAFI+YYNHACAEYSRQKM + FKL
Sbjct: 366 GVEDLKKIVTEIGPGVTGVELVKDMKNTNNNRGFAFIDYYNHACAEYSRQKMMSPTFKLG 545
Query: 178 NNAPTVSWAEPRNSESSAVSQVK 200
NAPTVSWA+P+N+ESSA SQVK
Sbjct: 546 ENAPTVSWADPKNAESSAASQVK 614
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/82 (34%), Positives = 41/82 (49%)
Frame = +3
Query: 202 VYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNT 261
VY+ +P +D LK L E G++ +V + K E FGFV F+ A KA++
Sbjct: 99 VYIGGIPHASDED-LKSLCERIGEVAEVRIMKGKXFFENKGFGFVTFTSVELASKAIEEL 275
Query: 262 EKYEIDGKNLECSLAKPEADQR 283
E GK ++CS K +A R
Sbjct: 276 NNTEFMGKKIKCS--KSQAKHR 335
>TC209876 similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A10_14, partial
(36%)
Length = 812
Score = 179 bits (454), Expect = 2e-45
Identities = 91/165 (55%), Positives = 126/165 (76%), Gaps = 3/165 (1%)
Frame = +1
Query: 165 SRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQVKAVYVKNLPENITQDSLKKLFEHH 223
SRQKM++S+FKL+ N PTV+WA+P+NS + SA SQVKA+YVKN+PEN+T + LK+LF H
Sbjct: 1 SRQKMASSSFKLDGNTPTVTWADPKNSPDHSASSQVKALYVKNIPENVTTEQLKELFRRH 180
Query: 224 GKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQR 283
G++TKVV+PP K+G ++ FGF+H+++RSSA+KA+K+TEKYEIDG+ LE LAKP+AD++
Sbjct: 181 GEVTKVVMPPGKAGGKRD-FGFIHYAERSSALKAVKDTEKYEIDGQMLEVVLAKPQADKK 357
Query: 284 SSGTSNSQKPVVLPTY-PHRLGYGMVGGAYGGIGAGYG-AAGFAQ 326
G + P + P + PH G YG +GAGYG AAG+ Q
Sbjct: 358 PDG-GYAYNPGLHPNHVPHPAYGNFSGNPYGSLGAGYGVAAGYQQ 489
>TC229796
Length = 413
Score = 163 bits (413), Expect = 1e-40
Identities = 79/110 (71%), Positives = 96/110 (86%), Gaps = 2/110 (1%)
Frame = +2
Query: 219 LFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKP 278
LFE HGKITKVVLPPAKSGQEK+R GFVHF++RS+AMKALKNTE+YE++G+ L+CSLAKP
Sbjct: 2 LFERHGKITKVVLPPAKSGQEKNRIGFVHFAERSNAMKALKNTERYELEGQLLQCSLAKP 181
Query: 279 EADQRSSGTSNSQK--PVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGFAQ 326
+ADQ+S G SN+QK P +LP+YP +GYG+VGGAYG +GAGY A G AQ
Sbjct: 182 QADQKSGG-SNTQKPGPGLLPSYPPHVGYGLVGGAYGALGAGYAAPGLAQ 328
>TC229795 weakly similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A10_14,
partial (12%)
Length = 652
Score = 95.9 bits (237), Expect = 3e-20
Identities = 46/74 (62%), Positives = 59/74 (79%), Gaps = 2/74 (2%)
Frame = +1
Query: 255 MKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPV--VLPTYPHRLGYGMVGGAY 312
MK K E+YE++G+ L+CSLAKP+ADQ+S G SN+QKP +LP+YP +GYG+VGGAY
Sbjct: 1 MKXXKXXERYELEGQLLQCSLAKPQADQKSGG-SNTQKPGPGLLPSYPPHVGYGLVGGAY 177
Query: 313 GGIGAGYGAAGFAQ 326
GG+GAGY A G AQ
Sbjct: 178 GGLGAGYAAPGLAQ 219
>TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(97%)
Length = 2392
Score = 87.4 bits (215), Expect = 9e-18
Identities = 65/269 (24%), Positives = 129/269 (47%), Gaps = 9/269 (3%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +Y+G + NV++ L +G+V VR+ + S GY +V F + A++A+
Sbjct: 177 TTSLYVGDLDPNVTDAQLYDLFNQLGQVVSVRVCRDLTSRRSLGYGYVNFSNPQDAARAL 356
Query: 85 EELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISI 136
+ LN + + I+ S + +FI N+ + + + + G ++S
Sbjct: 357 DVLNFTPLNNRPIRIMYSHRDPSIRKSGQGNIFIKNLDRAIDHKALHDTFSTFG-NILSC 533
Query: 137 ELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNSESSA- 195
++ D SSG+++G+ F+++ N A+ + +K+ N + + + ES+A
Sbjct: 534 KVATD--SSGQSKGYGFVQFDNEESAQKAIEKL-NGMLLNDKQVYVGPFLRKQERESAAD 704
Query: 196 VSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAM 255
++ V+VKNL E+ T D LK F G IT V+ G+ K FGFV+F + A
Sbjct: 705 KAKFNNVFVKNLSESTTDDELKNTFGEFGTITSAVVMRDGDGKSKC-FGFVNFENADDAA 881
Query: 256 KALKNTEKYEIDGKNLECSLAKPEADQRS 284
+A++ + D K A+ ++++ +
Sbjct: 882 RAVEALNGKKFDDKEWYVGKAQKKSEREN 968
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/80 (26%), Positives = 43/80 (53%)
Frame = +3
Query: 26 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIE 85
+ +Y+ + +++ +E L+ G ++ ++M+ +G +G FVAF T E AS+A+
Sbjct: 1026 ANLYVKNLDDSIGDEKLKELFSPFGTITSCKVMR-DPNGLSRGSGFVAFSTPEEASRALL 1202
Query: 86 ELNNSEFKGKKIKCSSSQAK 105
E+N K + + +Q K
Sbjct: 1203 EMNGKMVVSKPLYVTLAQRK 1262
Score = 35.8 bits (81), Expect = 0.032
Identities = 21/89 (23%), Positives = 41/89 (45%)
Frame = +3
Query: 201 AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKN 260
++YV +L N+T L LF G++ V + + + +G+V+FS+ A +AL
Sbjct: 183 SLYVGDLDPNVTDAQLYDLFNQLGQVVSVRVCRDLTSRRSLGYGYVNFSNPQDAARALDV 362
Query: 261 TEKYEIDGKNLECSLAKPEADQRSSGTSN 289
++ + + + + R SG N
Sbjct: 363 LNFTPLNNRPIRIMYSHRDPSIRKSGQGN 449
>TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (79%)
Length = 1560
Score = 83.2 bits (204), Expect = 2e-16
Identities = 71/328 (21%), Positives = 140/328 (42%), Gaps = 30/328 (9%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIE-- 85
++IG + + E L GEV+ V++++ K++ + +GY F+ F ++ A + ++
Sbjct: 312 LWIGDLQYWMDENYLYTCFAHTGEVTSVKVIRNKQTSQSEGYGFIEFNSRAGAERILQTY 491
Query: 86 ------------ELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGV 133
LN + F + + +F+G++ T +++ V
Sbjct: 492 NGAIMPNGGQSFRLNWATFSAGERSRHDDSPDYTIFVGDLAADVTDYLLQETFRARYNSV 671
Query: 134 ISIELLKDLQSSGRNRGFAFIEYYN-----HACAEYSRQKMSNSNFKL---ENNAPTVSW 185
+++ D + +GR +G+ F+ + + A E S ++ N PT
Sbjct: 672 KGAKVVID-RLTGRTKGYGFVRFSDESEQVRAMTEMQGVLCSTRPMRIGPASNKTPTTQS 848
Query: 186 --------AEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSG 237
++P+ S++ ++V NL N+T D L+++F +G++ V +P K
Sbjct: 849 QPKASYQNSQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIPAGK-- 1022
Query: 238 QEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLP 297
R GFV F+DRS A +AL+ + G+N+ S + +++++ N
Sbjct: 1023----RCGFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQADPNQWNG---- 1178
Query: 298 TYPHRLGYGMVGGAYGGIGAGYGAAGFA 325
G G GG YG GY G+A
Sbjct: 1179------GAGSGGGYYGYAAQGYENYGYA 1244
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/84 (23%), Positives = 42/84 (49%), Gaps = 4/84 (4%)
Frame = +3
Query: 180 APTVSWAE----PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAK 235
AP WA P + ++ +V+ +++ +L + ++ L F H G++T V + K
Sbjct: 234 APQAMWAPSAQPPPQQQPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVTSVKVIRNK 413
Query: 236 SGQEKSRFGFVHFSDRSSAMKALK 259
+ +GF+ F+ R+ A + L+
Sbjct: 414 QTSQSEGYGFIEFNSRAGAERILQ 485
>TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {Arabidopsis
thaliana;} , partial (64%)
Length = 1154
Score = 78.6 bits (192), Expect = 4e-15
Identities = 65/316 (20%), Positives = 138/316 (43%), Gaps = 30/316 (9%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIE-- 85
++IG + + E L GEVS V++++ K++ + +GY F+ F ++ A + ++
Sbjct: 246 LWIGDLQYWMDENYLYTCFAHTGEVSSVKVIRNKQTSQSEGYGFIEFNSRAGAERILQTY 425
Query: 86 ------------ELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGV 133
LN + F + + +F+G++ T +++ V
Sbjct: 426 NGAIMPNGGQSFRLNWATFSAGERSRQDDSPDYTIFVGDLAADVTDYLLQETFRARYNSV 605
Query: 134 ISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKM--------------SNSNFKLENN 179
+++ D + +GR +G+ F+ + + + +M +++ +
Sbjct: 606 KGAKVVID-RLTGRTKGYGFVRFSEESEQMRAMTEMQGVLCSTRPMRIGPASNKTPATQS 782
Query: 180 APTVSW--AEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSG 237
P S+ ++P+ S++ ++V NL N+T D L+++F +G++ V +P K
Sbjct: 783 QPKASYLNSQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIPAGK-- 956
Query: 238 QEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLP 297
R GFV F+DRS A +AL+ + G+N+ S + +++++ N
Sbjct: 957 ----RCGFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQADPNQ------- 1103
Query: 298 TYPHRLGYGMVGGAYG 313
+ G G GG YG
Sbjct: 1104-WNGAAGAGSGGGYYG 1148
Score = 37.7 bits (86), Expect = 0.008
Identities = 19/87 (21%), Positives = 44/87 (49%), Gaps = 4/87 (4%)
Frame = +3
Query: 177 ENNAPTVSWAE----PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLP 232
++ AP WA P + ++ +V+ +++ +L + ++ L F H G+++ V +
Sbjct: 159 QHQAPQPMWAPSAQPPLPQQPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVSSVKVI 338
Query: 233 PAKSGQEKSRFGFVHFSDRSSAMKALK 259
K + +GF+ F+ R+ A + L+
Sbjct: 339 RNKQTSQSEGYGFIEFNSRAGAERILQ 419
>TC225576 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial
(93%)
Length = 2491
Score = 73.6 bits (179), Expect = 1e-13
Identities = 56/216 (25%), Positives = 102/216 (46%), Gaps = 10/216 (4%)
Frame = +3
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRL--------FIGNIPKKWTV 119
GY +V F + A++A++ LN + + I+ S L FI N+ K
Sbjct: 486 GYGYVNFSNPQDAARALDVLNFTPLNNRPIRIMYSHRDPSLRKSGTANIFIKNLDKAIDH 665
Query: 120 EDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENN 179
+ + + G ++S ++ D +SG ++G+ F+++ N A+ + K++ + +
Sbjct: 666 KALHDTFSSFGL-ILSCKIATD--ASGLSKGYGFVQFDNEEAAQNAIDKLNG--MLINDK 830
Query: 180 APTVSWAEPRNSESSAVSQVKA--VYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSG 237
V + +A+S+ K VYVKNL E+ T + L K F +G IT V+ G
Sbjct: 831 QVYVGHFLRKQDRENALSKTKFNNVYVKNLSESTTDEELMKFFGEYGTITSAVIMRDADG 1010
Query: 238 QEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLEC 273
+ + FGFV+F + A KA++ ++D K C
Sbjct: 1011KSRC-FGFVNFENPDDAAKAVEGLNGKKVDDKEWXC 1115
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/78 (30%), Positives = 44/78 (55%)
Frame = +1
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
+Y+ + + +S+E L+ G ++ ++M+ +G +G FVAF T E AS+A+ E+
Sbjct: 1213 LYLKNLDDTISDEKLKEMFAEYGTITSCKVMR-DPTGIGRGSGFVAFSTPEEASRALGEM 1389
Query: 88 NNSEFKGKKIKCSSSQAK 105
N F GK + + +Q K
Sbjct: 1390 NGKMFAGKPLYVALAQRK 1443
>TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding protein, partial
(70%)
Length = 1724
Score = 72.4 bits (176), Expect = 3e-13
Identities = 65/285 (22%), Positives = 130/285 (44%), Gaps = 29/285 (10%)
Frame = +1
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIE-- 85
++IG + + + E L S GE+S +++++ K++G +GY FV F + A + ++
Sbjct: 430 IWIGDLHHWMDENYLHRCFASTGEISSIKVIRNKQTGLSEGYGFVEFYSHATAEKVLQNY 609
Query: 86 ------------ELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGV 133
LN + F K S + +F+G++ T + + A V P V
Sbjct: 610 AGILMPNAEQPFRLNWATFSTGD-KGSDNVPDLSIFVGDLAADVTDSLLHETFASVYPSV 786
Query: 134 ISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAP-TVSWAEPRNS- 191
+ +++ D ++GR++G+ F+ + + Q M+ N ++ P + A PR S
Sbjct: 787 KAAKVVFD-ANTGRSKGYGFVRFGDD---NERTQAMTQMNGVYCSSRPMRIGAATPRKSS 954
Query: 192 -------------ESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQ 238
+S A S ++V L N++ + L++ F +G+I V +P K
Sbjct: 955 GHQQGGLSNGTANQSEADSTNTTIFVGGLDPNVSDEDLRQPFSQYGEIVSVKIPVGKG-- 1128
Query: 239 EKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQR 283
GFV F++R++A +AL+ I + + S + A+++
Sbjct: 1129----CGFVQFANRNNAEEALQKLNGTTIGKQTVRLSWGRNPANKQ 1251
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/76 (30%), Positives = 41/76 (53%)
Frame = +1
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +++GG+ NVS+EDLR GE+ V+I KG FV F + A +A+
Sbjct: 1015 NTTIFVGGLDPNVSDEDLRQPFSQYGEIVSVKIPVG------KGCGFVQFANRNNAEEAL 1176
Query: 85 EELNNSEFKGKKIKCS 100
++LN + + ++ S
Sbjct: 1177 QKLNGTTIGKQTVRLS 1224
>TC217407 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1194
Score = 65.9 bits (159), Expect = 3e-11
Identities = 47/185 (25%), Positives = 83/185 (44%), Gaps = 8/185 (4%)
Frame = +2
Query: 100 SSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNH 159
++ A RL++GNIP+ T +++ K+V + G V E++ D + SGR+R FAF+
Sbjct: 278 ATEAAARRLYVGNIPRTVTNDELAKIVQEHG-AVEKAEVMYD-KYSGRSRRFAFVTMKTV 451
Query: 160 ACAEYSRQKMSNSNFK--------LENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENI 211
A +K++ + E T+ + ES + VYV NL + +
Sbjct: 452 EDATAVIEKLNGTELGGREIKVNVTEKPLSTLDLPLLQAEESEFIDSPHKVYVGNLAKTV 631
Query: 212 TQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNL 271
T D+LK F GK+ + + S +GFV F A+ + ++G+ +
Sbjct: 632 TTDTLKNFFSEKGKVLSAKVSRVPGTSKSSGYGFVTFPSEEDVEAAISSFNNSLLEGQTI 811
Query: 272 ECSLA 276
+ A
Sbjct: 812 RVNKA 826
Score = 49.7 bits (117), Expect = 2e-06
Identities = 24/80 (30%), Positives = 43/80 (53%)
Frame = +2
Query: 23 PHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQ 82
PH +VY+G +A V+ + L+ F G+V ++ + + + GY FV F ++E
Sbjct: 593 PH--KVYVGNLAKTVTTDTLKNFFSEKGKVLSAKVSRVPGTSKSSGYGFVTFPSEEDVEA 766
Query: 83 AIEELNNSEFKGKKIKCSSS 102
AI NNS +G+ I+ + +
Sbjct: 767 AISSFNNSLLEGQTIRVNKA 826
Score = 42.4 bits (98), Expect = 3e-04
Identities = 20/92 (21%), Positives = 41/92 (43%)
Frame = +2
Query: 186 AEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGF 245
A + ++ + + +YV N+P +T D L K+ + HG + K + K RF F
Sbjct: 254 AVAEQAATATEAAARRLYVGNIPRTVTNDELAKIVQEHGAVEKAEVMYDKYSGRSRRFAF 433
Query: 246 VHFSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
V A ++ E+ G+ ++ ++ +
Sbjct: 434 VTMKTVEDATAVIEKLNGTELGGREIKVNVTE 529
>TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (85%)
Length = 1436
Score = 65.1 bits (157), Expect = 5e-11
Identities = 44/178 (24%), Positives = 86/178 (47%), Gaps = 11/178 (6%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++++Y G + +V L Q G + ++ +++G+ +G+AFV E + I
Sbjct: 495 ATKLYFGNLPYSVDSAKLAGLIQDYGSAELIEVLYDRDTGKSRGFAFVTMSCIEDCNAVI 674
Query: 85 EELNNSEFKGKKIKCSSS-----------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGV 133
E L+ EF G+ ++ + S + +H+LF+GN+ T E + + + G V
Sbjct: 675 ENLDGKEFLGRTLRVNFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGT-V 851
Query: 134 ISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS 191
+ +L D +GR+RG+ F+ Y AE + ++ +LE A VS A+ + +
Sbjct: 852 VGARVLYD-GETGRSRGYGFVCYSTK--AEMEAALAALNDVELEGRAMRVSLAQGKRA 1016
Score = 57.0 bits (136), Expect = 1e-08
Identities = 49/198 (24%), Positives = 96/198 (47%), Gaps = 4/198 (2%)
Frame = +3
Query: 84 IEELNNSEFKGKKI-KCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDL 142
+EE E +G+ + + S + +L+ GN+P + ++ D G + IE+L D
Sbjct: 429 VEEEKVEENEGEAVAEQDSDSSATKLYFGNLPYSVDSAKLAGLIQDYGSAEL-IEVLYD- 602
Query: 143 QSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFK--LENNAPTVSWAEPRNSESSAVSQVK 200
+ +G++RGFAF+ +C E + N + K L ++P+ E
Sbjct: 603 RDTGKSRGFAFVTM---SCIEDCNAVIENLDGKEFLGRTLRVNFSSKPKPKEPLYPETEH 773
Query: 201 AVYVKNLPENITQDSLKKLFEHHGKITKV-VLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
++V NL ++T + L + F+ +G + VL ++G+ + +GFV +S ++ AL
Sbjct: 774 KLFVGNLSWSVTNEILTQAFQEYGTVVGARVLYDGETGRSRG-YGFVCYSTKAEMEAALA 950
Query: 260 NTEKYEIDGKNLECSLAK 277
E++G+ + SLA+
Sbjct: 951 ALNDVELEGRAMRVSLAQ 1004
>TC214546 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (68%)
Length = 1756
Score = 64.3 bits (155), Expect = 8e-11
Identities = 66/338 (19%), Positives = 135/338 (39%), Gaps = 40/338 (11%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEE- 86
V++G + + + E L GEV ++++ K++G+ +GY FV F ++ A + ++
Sbjct: 495 VWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRATAEKVLQNY 674
Query: 87 -------------LNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGV 133
LN + F + + S + + +F+G++ T +++ A +
Sbjct: 675 NGTMMPNTDQAFRLNWATFSAGERRSSDATSDLSIFVGDLAIDVTDAMLQETFAGRYSSI 854
Query: 134 ISIELLKDLQSSGRNRGFAFIEYYNHA-------------CAE-------------YSRQ 167
+++ D ++GR++G+ F+ + + C+ Y Q
Sbjct: 855 KGAKVVID-SNTGRSKGYGFVRFGDENERTRAMTEMNGVYCSSRPMRIGVATPKKTYGFQ 1031
Query: 168 KMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKIT 227
+ +S + + + A + S S ++V L + + + L++ F G++
Sbjct: 1032QQYSSQAVVLAGGHSANGAVAQGSHSEGDINNTTIFVGGLDSDTSDEDLRQPFLQFGEVV 1211
Query: 228 KVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGT 287
V +P K GFV F+DR +A +A++ I + + S + ++
Sbjct: 1212SVKIPVGKG------CGFVQFADRKNAEEAIQGLNGTVIGKQTVRLSWGRSPGNKHWRSD 1373
Query: 288 SNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGFA 325
SN GG YGG GYG GFA
Sbjct: 1374SN-------------------GGHYGG-HQGYGGHGFA 1427
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/77 (29%), Positives = 42/77 (53%)
Frame = +3
Query: 24 HSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQA 83
+++ +++GG+ ++ S+EDLR GEV V+I KG FV F ++ A +A
Sbjct: 1122 NNTTIFVGGLDSDTSDEDLRQPFLQFGEVVSVKIPVG------KGCGFVQFADRKNAEEA 1283
Query: 84 IEELNNSEFKGKKIKCS 100
I+ LN + + ++ S
Sbjct: 1284 IQGLNGTVIGKQTVRLS 1334
Score = 34.3 bits (77), Expect = 0.092
Identities = 16/68 (23%), Positives = 36/68 (52%)
Frame = +3
Query: 193 SSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRS 252
+++ +++ V++ +L + ++ L F H G++ + K + +GFV F R+
Sbjct: 468 AASSDEIRTVWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRA 647
Query: 253 SAMKALKN 260
+A K L+N
Sbjct: 648 TAEKVLQN 671
>TC225236 similar to UP|Q84LL7 (Q84LL7) Salt tolerance protein 6, partial
(39%)
Length = 825
Score = 63.5 bits (153), Expect = 1e-10
Identities = 39/140 (27%), Positives = 68/140 (47%)
Frame = +3
Query: 186 AEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGF 245
++P+ S++ ++V NL N+T D L+++F +G++ V +P K R GF
Sbjct: 153 SQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIPAGK------RCGF 314
Query: 246 VHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGY 305
V F+DRS A +AL+ + G+N+ S + +++++ N + G
Sbjct: 315 VQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQADPNQ--------WNGAAGA 470
Query: 306 GMVGGAYGGIGAGYGAAGFA 325
G GG YG GY G+A
Sbjct: 471 GSGGGYYGYAAQGYENYGYA 530
>TC204822 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (67%)
Length = 1232
Score = 62.8 bits (151), Expect = 2e-10
Identities = 45/178 (25%), Positives = 87/178 (48%), Gaps = 7/178 (3%)
Frame = +3
Query: 107 RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSR 166
+LF+GN+P + V+ K + G + I + + + ++RGF F+ AE +
Sbjct: 333 KLFVGNLP--YDVDSQKLAMLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSTVEEAESAV 506
Query: 167 QKMSNSNFKLENNAPTVSWAEPRNSESS------AVSQVKAVYVKNLPENITQDSLKKLF 220
+K + + ++ TV+ A PR + + ++YV NLP ++ LK++F
Sbjct: 507 EKFNR--YDIDGRLLTVNKASPRGTRPERPPPRRSFESSLSIYVGNLPWDVDNTRLKQIF 680
Query: 221 EHHGKITKV-VLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
HG + V+ +SG+ + FGFV SD + A+ + +DG+ ++ S+A+
Sbjct: 681 SKHGNVVNARVVYDRESGRSRG-FGFVTMSDETEMNDAVAALDGESLDGRAIKVSVAE 851
Score = 44.7 bits (104), Expect = 7e-05
Identities = 31/107 (28%), Positives = 48/107 (43%), Gaps = 5/107 (4%)
Frame = +3
Query: 177 ENNAPTVSWAEPRNSESSAVSQVKA-----VYVKNLPENITQDSLKKLFEHHGKITKVVL 231
E A SW EP ++ S V+ ++V NLP ++ L LFE G + +
Sbjct: 249 ETEAGLESW-EPNGEDAGDESFVEPPEEAKLFVGNLPYDVDSQKLAMLFEQAGTVEIAEV 425
Query: 232 PPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKP 278
+ + FGFV S A A++ +Y+IDG+ L + A P
Sbjct: 426 IYNRETDQSRGFGFVTMSTVEEAESAVEKFNRYDIDGRLLTVNKASP 566
Score = 42.0 bits (97), Expect = 4e-04
Identities = 21/83 (25%), Positives = 39/83 (46%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
S +Y+G + +V L+ G V R++ +ESG +G+ FV + + A+
Sbjct: 615 SLSIYVGNLPWDVDNTRLKQIFSKHGNVVNARVVYDRESGRSRGFGFVTMSDETEMNDAV 794
Query: 85 EELNNSEFKGKKIKCSSSQAKHR 107
L+ G+ IK S ++ + R
Sbjct: 795 AALDGESLDGRAIKVSVAEDRPR 863
>TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45,
partial (55%)
Length = 886
Score = 62.4 bits (150), Expect = 3e-10
Identities = 60/268 (22%), Positives = 115/268 (42%), Gaps = 33/268 (12%)
Frame = +1
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
++IG + V E L GEV ++I++ K +G+ +GY FV F + A +
Sbjct: 70 LWIGDLQYWVDESYLSQCFAHNGEVVSIKIIRNKLTGQPEGYGFVEFVSHASAEAFLRTY 249
Query: 88 NNSEFKGKK-------IKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
N ++ G + S H +F+G++ T +++ P V +++
Sbjct: 250 NGAQMPGTEQTFRLNWASFGDSGPDHSIFVGDLAPDVTDFLLQETFRAHYPSVKGAKVVT 429
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMS------------------NSNFKLENNAPT 182
D ++GR++G+ F+++ + A + +M+ N++F+ + P
Sbjct: 430 D-PATGRSKGYGFVKFADEAQRNRAMTEMNGVYCSTRPMRISAATPKKNASFQHQYAPPK 606
Query: 183 VSWAEPRNSESSAVSQV--------KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPA 234
+ P + S+ VS V V + NL N+T++ LK+ F G I V +
Sbjct: 607 AMYQFP--AYSAPVSAVAPENDVNNTTVCIGNLDLNVTEEELKQTFMQFGDIVLVKIYAG 780
Query: 235 KSGQEKSRFGFVHFSDRSSAMKALKNTE 262
K +G+V F R SA A++ +
Sbjct: 781 KG------YGYVQFGTRVSAEDAIQRMQ 846
Score = 49.7 bits (117), Expect = 2e-06
Identities = 52/217 (23%), Positives = 99/217 (44%), Gaps = 3/217 (1%)
Frame = +1
Query: 66 RKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKV 125
+KG A+ A + K +A+ AIEE+ L+IG++ + W E
Sbjct: 1 KKGEAYSARRWKMVAT-AIEEVRT------------------LWIGDL-QYWVDESYLSQ 120
Query: 126 VADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSW 185
V+SI+++++ + +G+ G+ F+E+ +HA AE + + + ++W
Sbjct: 121 CFAHNGEVVSIKIIRN-KLTGQPEGYGFVEFVSHASAEAFLRTYNGAQMPGTEQTFRLNW 297
Query: 186 AEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHG---KITKVVLPPAKSGQEKSR 242
A +S +++V +L ++T L++ F H K KVV PA +G+ K
Sbjct: 298 ASFGDSGPD-----HSIFVGDLAPDVTDFLLQETFRAHYPSVKGAKVVTDPA-TGRSKG- 456
Query: 243 FGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPE 279
+GFV F+D + +A+ + + S A P+
Sbjct: 457 YGFVKFADEAQRNRAMTEMNGVYCSTRPMRISAATPK 567
>TC225238 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (31%)
Length = 1258
Score = 62.4 bits (150), Expect = 3e-10
Identities = 39/142 (27%), Positives = 68/142 (47%)
Frame = +1
Query: 184 SWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRF 243
S+ P+ +++ ++V NL N+T D L+++F H+G++ V +P K R
Sbjct: 583 SYQNPQGAQNEHDPNNTTIFVGNLDPNVTDDHLRQVFGHYGELVHVKIPAGK------RC 744
Query: 244 GFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRL 303
GFV F+DRS A +AL+ + G+N+ S + +++++ +N
Sbjct: 745 GFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQPDANQWN----------- 891
Query: 304 GYGMVGGAYGGIGAGYGAAGFA 325
G GG YG GY G+A
Sbjct: 892 --GSGGGYYGYAQGGYENYGYA 951
>TC225240 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (48%)
Length = 928
Score = 62.0 bits (149), Expect = 4e-10
Identities = 48/196 (24%), Positives = 88/196 (44%), Gaps = 15/196 (7%)
Frame = +1
Query: 145 SGRNRGFAFIEYYNHACAEYSRQKM---------------SNSNFKLENNAPTVSWAEPR 189
+GR +G+ F+ + + + + +M SN N ++ P S+ P+
Sbjct: 58 TGRTKGYGFVRFGDESEQVRAMTEMQGVLCSTRPMRIGPASNKNPSTQSQ-PKASYQNPQ 234
Query: 190 NSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFS 249
+++ ++V NL N+T D L+++F +G++ V +P K R GFV F+
Sbjct: 235 GAQNEHDPNNTTIFVGNLDPNVTDDHLRQVFGQYGELVHVKIPAGK------RCGFVQFA 396
Query: 250 DRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVG 309
DRS A +AL+ + G+N+ S + +++++ +N G G G
Sbjct: 397 DRSCAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQPDANQWN-----------GSG-GG 540
Query: 310 GAYGGIGAGYGAAGFA 325
G YG GY G+A
Sbjct: 541 GYYGYAQGGYENYGYA 588
>TC225741 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (57%)
Length = 857
Score = 60.5 bits (145), Expect = 1e-09
Identities = 43/167 (25%), Positives = 79/167 (46%), Gaps = 11/167 (6%)
Frame = +2
Query: 36 NVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELNNSEFKGK 95
+V L Q G + ++ ++SG+ +G+AFV E + IE L+ EF G+
Sbjct: 2 SVDSXKLAGLIQDYGSAELIEVLYDRDSGKSRGFAFVTMSCIEDCNAVIENLDGKEFLGR 181
Query: 96 KIKCSSS-----------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQS 144
++ + S + +H+LF+GN+ T E + + + G V+ +L D
Sbjct: 182 XLRVNFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGT-VVGARVLYD-GE 355
Query: 145 SGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS 191
+GR+RG+ F+ Y AE + ++ +LE A VS A+ + +
Sbjct: 356 TGRSRGYGFVCYSTQ--AEMEAAVAALNDVELEGRAMRVSLAQGKRA 490
Score = 46.2 bits (108), Expect = 2e-05
Identities = 40/156 (25%), Positives = 77/156 (48%), Gaps = 3/156 (1%)
Frame = +2
Query: 125 VVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFK--LENNAPT 182
++ D G + IE+L D + SG++RGFAF+ +C E + N + K L
Sbjct: 29 LIQDYGSAEL-IEVLYD-RDSGKSRGFAFVTM---SCIEDCNAVIENLDGKEFLGRXLRV 193
Query: 183 VSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKV-VLPPAKSGQEKS 241
++P+ E ++V NL ++T + L + F+ +G + VL ++G+ +
Sbjct: 194 NFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGTVVGARVLYDGETGRSRG 373
Query: 242 RFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
+GFV +S ++ A+ E++G+ + SLA+
Sbjct: 374 -YGFVCYSTQAEMEAAVAALNDVELEGRAMRVSLAQ 478
>TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, partial (9%)
Length = 1513
Score = 60.1 bits (144), Expect = 2e-09
Identities = 56/253 (22%), Positives = 112/253 (44%), Gaps = 40/253 (15%)
Frame = +3
Query: 50 GEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELNNSEFK-------------GKK 96
GEV ++I++ K +G+ +GY FV F + A + ++ N ++ G
Sbjct: 39 GEVISIKIIRNKLTGQPEGYGFVEFVSHAAAERVLQTYNGTQMPATDQTFRLNWASFGIG 218
Query: 97 IKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEY 156
+ + +H +F+G++ T +++ P V +++ D ++ R++G+ F+++
Sbjct: 219 ERRPDAAPEHSIFVGDLAPDVTDYLLQETFRAHYPSVRGAKVVTD-PNTARSKGYGFVKF 395
Query: 157 -----YNHACAEYSRQKMSNSNFKLENNAP---TVSWAEPR----------NSESSAVSQ 198
N A E + S ++ P T ++A P + +S V Q
Sbjct: 396 SDENERNRAMTEMNGVYCSTRPMRISAATPKKTTGAYAAPAAPVPKPVYPVPAYTSPVVQ 575
Query: 199 VK---------AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFS 249
V+ ++V NL N++++ LK+ G+I V + P K FGFV F
Sbjct: 576 VQPPDYDVNNTTIFVGNLDLNVSEEELKQNSLQFGEIVSVKIQPGKG------FGFVQFG 737
Query: 250 DRSSAMKALKNTE 262
R+SA +A++ +
Sbjct: 738 TRASAEEAIQKMQ 776
Score = 51.2 bits (121), Expect = 7e-07
Identities = 41/172 (23%), Positives = 79/172 (45%), Gaps = 4/172 (2%)
Frame = +3
Query: 133 VISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNSE 192
VISI+++++ + +G+ G+ F+E+ +HA AE Q + + + ++WA E
Sbjct: 45 VISIKIIRN-KLTGQPEGYGFVEFVSHAAAERVLQTYNGTQMPATDQTFRLNWASFGIGE 221
Query: 193 SSA-VSQVKAVYVKNLPENITQDSLKKLFEHHG---KITKVVLPPAKSGQEKSRFGFVHF 248
+ +++V +L ++T L++ F H + KVV P + +GFV F
Sbjct: 222 RRPDAAPEHSIFVGDLAPDVTDYLLQETFRAHYPSVRGAKVVTDPNTA--RSKGYGFVKF 395
Query: 249 SDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYP 300
SD + +A+ + + S A P+ + + + PV P YP
Sbjct: 396 SDENERNRAMTEMNGVYCSTRPMRISAATPK--KTTGAYAAPAAPVPKPVYP 545
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/77 (27%), Positives = 41/77 (52%)
Frame = +3
Query: 24 HSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQA 83
+++ +++G + NVSEE+L+ GE+ V+I KG+ FV F T+ A +A
Sbjct: 600 NNTTIFVGNLDLNVSEEELKQNSLQFGEIVSVKIQPG------KGFGFVQFGTRASAEEA 761
Query: 84 IEELNNSEFKGKKIKCS 100
I+++ + ++ S
Sbjct: 762 IQKMQGKMIGQQVVRIS 812
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,673,897
Number of Sequences: 63676
Number of extensions: 227368
Number of successful extensions: 2098
Number of sequences better than 10.0: 214
Number of HSP's better than 10.0 without gapping: 1904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2054
length of query: 370
length of database: 12,639,632
effective HSP length: 99
effective length of query: 271
effective length of database: 6,335,708
effective search space: 1716976868
effective search space used: 1716976868
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0010.2


