

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
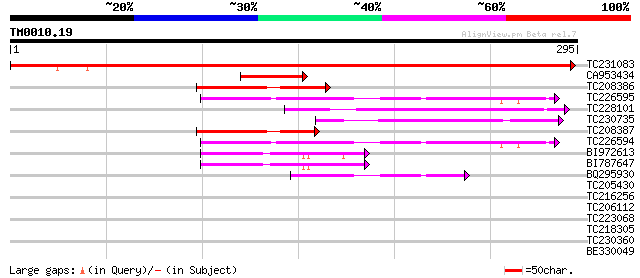
Query= TM0010.19
(295 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231083 460 e-130
CA953434 64 1e-10
TC208386 63 1e-10
TC226595 similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partia... 58 6e-09
TC228101 56 2e-08
TC230735 54 6e-08
TC208387 54 1e-07
TC226594 weakly similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein,... 53 1e-07
BI972613 52 3e-07
BI787647 49 2e-06
BQ295930 similar to GP|21592576|gb| SOUL-like protein {Arabidops... 43 1e-04
TC205430 similar to UP|O22600 (O22600) Glycine-rich protein, par... 30 1.3
TC216256 homologue to UP|Q6PQQ3 (Q6PQQ3) AP2/EREBP transcription... 30 1.7
TC206112 similar to UP|Q8VIL5 (Q8VIL5) Ski proto-oncogene, parti... 28 3.8
TC223068 28 4.9
TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like p... 28 4.9
TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {A... 28 6.4
BE330049 homologue to GP|2252828|gb| A_IG005I10.5 gene product {... 27 8.4
>TC231083
Length = 1086
Score = 460 bits (1184), Expect = e-130
Identities = 239/302 (79%), Positives = 264/302 (87%), Gaps = 8/302 (2%)
Frame = +1
Query: 1 MLPLCN-ASLSSQSSIRTSIPGRP-----NIAITNSASSSERISN--RRRTMSAVEARSS 52
M +CN +SLS Q I IP +P N+++TNSASS ERIS+ RRRT+S E+R S
Sbjct: 79 MFTICNPSSLSRQPGIVAPIPVKPIKHSSNLSVTNSASSGERISSSTRRRTISPFESRIS 258
Query: 53 LILALASQANSLTQRLVVDVATETARYLFPKRLESRTLEEALMAVPDLETVKFKVLSRRD 112
LI ALASQANSL+QRL+ DVA ETA+Y+FPKR ESR LEEALM+VPDLETV FKVLSR D
Sbjct: 259 LIFALASQANSLSQRLLADVAAETAKYVFPKRFESRNLEEALMSVPDLETVDFKVLSRMD 438
Query: 113 NYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKN 172
YEIREVEPYFVAE TMPGKSGFDF+GAS+SFN LAEYLFGKNT KEKMEMTTPV+TSKN
Sbjct: 439 QYEIREVEPYFVAETTMPGKSGFDFNGASRSFNALAEYLFGKNTTKEKMEMTTPVFTSKN 618
Query: 173 QSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRKIVAVVS 232
QSDGVKMDMTTPV T KMEDQD W MSF++PSKYGANLPLPKDSSVRIKE+PRKIVAVVS
Sbjct: 619 QSDGVKMDMTTPVLTTKMEDQDNWKMSFVMPSKYGANLPLPKDSSVRIKEVPRKIVAVVS 798
Query: 233 FSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEIALEVER 292
FSGFVNDEE+KQRELKLR+ALKSD QF+IK+GTSVEVAQYNPPFTLPFQRRNEIALEVE
Sbjct: 799 FSGFVNDEEIKQRELKLRDALKSDSQFEIKEGTSVEVAQYNPPFTLPFQRRNEIALEVEW 978
Query: 293 KD 294
K+
Sbjct: 979 KN 984
>CA953434
Length = 427
Score = 63.5 bits (153), Expect = 1e-10
Identities = 29/35 (82%), Positives = 31/35 (87%)
Frame = +2
Query: 121 PYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKN 155
PYFVAE TMPGKSGFDF+GAS+SFN LAEYLF N
Sbjct: 308 PYFVAETTMPGKSGFDFNGASRSFNALAEYLFEYN 412
>TC208386
Length = 757
Score = 63.2 bits (152), Expect = 1e-10
Identities = 31/70 (44%), Positives = 44/70 (62%)
Frame = +1
Query: 98 PDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTL 157
P+LE+ K+++L R +NYE+R+ P+ V E SG S FN +A Y+FGKN+
Sbjct: 565 PELESPKYQILKRTENYEVRQYNPFIVVETNGDKLSG------STGFNDVAGYIFGKNST 726
Query: 158 KEKMEMTTPV 167
EK+ MTTPV
Sbjct: 727 TEKIPMTTPV 756
>TC226595 similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partial (59%)
Length = 950
Score = 57.8 bits (138), Expect = 6e-09
Identities = 55/196 (28%), Positives = 86/196 (43%), Gaps = 9/196 (4%)
Frame = +1
Query: 100 LETVKFKVLSRRDNYEIREVE-PYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLK 158
+E + V+ + YEIR P +++ + + S + F L +Y+ GKN K
Sbjct: 193 IECPSYDVIHVGNGYEIRRYNSPVWISNSPIQDISLVE--ATRTGFRRLFDYIQGKNNYK 366
Query: 159 EKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSV 218
+K+EMT PV + SDG + + +SF +P + AN P K +
Sbjct: 367 QKIEMTAPVISEVLPSDGPFC-------------ESSFVVSFYVPKENQANPPPAK--GL 501
Query: 219 RIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALK----SDGQFKIKK----GTSVEVA 270
+++ VAV F GFV D V + L+ ++ +D K +K + VA
Sbjct: 502 QVQRWKTVFVAVRQFGGFVKDSSVGEEAAALKASIAGTKWADAVEKSQKRAGHASVYTVA 681
Query: 271 QYNPPFTLPFQRRNEI 286
QYN PF R NEI
Sbjct: 682 QYNAPFEYD-NRVNEI 726
>TC228101
Length = 952
Score = 55.8 bits (133), Expect = 2e-08
Identities = 41/149 (27%), Positives = 69/149 (45%), Gaps = 1/149 (0%)
Frame = +3
Query: 144 FNVLAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILP 203
F+ L ++ G N ++ MT PV +TT V A + +S LP
Sbjct: 207 FHRLFQFTEGANLNFSRIPMTIPV-------------LTTAVPGAGPLQSQGYYVSLYLP 347
Query: 204 SKYGANLPLP-KDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIK 262
K+ + P+P + +++ E VAV FSGF DE + + KL +L + K
Sbjct: 348 VKFQGDPPVPLPELNIKPYEFSSHCVAVRKFSGFAKDERIVKEAEKLATSLSRSPWAESK 527
Query: 263 KGTSVEVAQYNPPFTLPFQRRNEIALEVE 291
G +AQYN P + +R+NE+ ++++
Sbjct: 528 TGRGYSIAQYNTPIRI-VKRKNEVWVDID 611
>TC230735
Length = 612
Score = 54.3 bits (129), Expect = 6e-08
Identities = 39/131 (29%), Positives = 65/131 (48%), Gaps = 2/131 (1%)
Frame = +3
Query: 160 KMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILP-SKYGANLPLPKDSSV 218
++ MTTPV+T N + D K ++ +LP K +LP P +V
Sbjct: 12 EIPMTTPVFTETNDA-----------------DLSKVSIQIVLPLDKETESLPNPNQETV 140
Query: 219 RIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEVAQYNPP-FT 277
R++++ I AV+ FSG ++ V+++E LR + DG +K + +A+YN P T
Sbjct: 141 RLRKVEGGIAAVMKFSGKPTEDTVREKEKTLRANIIKDG---LKPQSGCLLARYNDPGRT 311
Query: 278 LPFQRRNEIAL 288
F RNE+ +
Sbjct: 312 WTFIMRNEVLI 344
>TC208387
Length = 487
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/64 (40%), Positives = 39/64 (60%)
Frame = +2
Query: 98 PDLETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTL 157
P+LE+ K+++L R +NYE+R+ P+ V E SG S FN +A Y+FGKN+
Sbjct: 311 PELESPKYQILKRTENYEVRQYNPFIVVETNGDKLSG------STGFNDVAGYIFGKNST 472
Query: 158 KEKM 161
EK+
Sbjct: 473 TEKI 484
>TC226594 weakly similar to UP|Q9SHG8 (Q9SHG8) SOUL-like protein, partial
(77%)
Length = 901
Score = 53.1 bits (126), Expect = 1e-07
Identities = 53/196 (27%), Positives = 83/196 (42%), Gaps = 9/196 (4%)
Frame = +3
Query: 100 LETVKFKVLSRRDNYEIREVE-PYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLK 158
+E + V+ + YEIR P +++ + + S + F L +Y+ GKN K
Sbjct: 84 IECPSYDVIHFGNGYEIRRYNSPVWISNSPILDISLVE--ATRTGFRRLFDYIQGKNNYK 257
Query: 159 EKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSV 218
+K+EMT PV + DG + + +SF +P + AN P K +
Sbjct: 258 QKIEMTAPVISEVLPRDGPFC-------------ESSFVVSFYVPKENQANPPPAK--GL 392
Query: 219 RIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALK----SDGQFKIKK----GTSVEVA 270
++ AV F GFV D V + L+ ++ +D K +K + VA
Sbjct: 393 HVQRWKTVFAAVRQFGGFVKDSSVGEEAAALKASIAGTKWADAVEKSQKRAGHASVYTVA 572
Query: 271 QYNPPFTLPFQRRNEI 286
QYN PF R NEI
Sbjct: 573 QYNAPFEYD-NRVNEI 617
>BI972613
Length = 423
Score = 52.0 bits (123), Expect = 3e-07
Identities = 39/97 (40%), Positives = 49/97 (50%), Gaps = 9/97 (9%)
Frame = +2
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQ-SFNVLAEYL--FGK-- 154
+ET K++V+ YEIR+ P VAE T F G F +LA Y+ GK
Sbjct: 131 VETAKYEVIKSTSEYEIRKYAPSVVAEVTYDPS---QFKGNKDGGFMILANYIGAVGKPQ 301
Query: 155 NTLKEKMEMTTPVYTSKN----QSDGVKMDMTTPVYT 187
NT EK+ MT PV T + SDG K+ MT PV T
Sbjct: 302 NTKPEKIAMTAPVITKDSVGGGGSDGEKIAMTAPVVT 412
>BI787647
Length = 423
Score = 49.3 bits (116), Expect = 2e-06
Identities = 37/94 (39%), Positives = 47/94 (49%), Gaps = 6/94 (6%)
Frame = +2
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQ-SFNVLAEYL--FGK-- 154
+ET K++V+ YEIR+ P VAE T F G F +LA Y+ GK
Sbjct: 146 VETAKYEVIKSTSEYEIRKYAPSVVAEVTYDPS---QFKGNKDGGFMILANYIGAVGKPQ 316
Query: 155 NTLKEKMEMTTPVYTSKN-QSDGVKMDMTTPVYT 187
NT EK+ MT PV T + DG + MT PV T
Sbjct: 317 NTKPEKIAMTAPVITKDSVGGDGETIAMTAPVVT 418
>BQ295930 similar to GP|21592576|gb| SOUL-like protein {Arabidopsis
thaliana}, partial (40%)
Length = 420
Score = 43.1 bits (100), Expect = 1e-04
Identities = 30/93 (32%), Positives = 42/93 (44%)
Frame = +1
Query: 147 LAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKY 206
L +Y+ GKN K+K+EMT PV T + SDG + + +SF +P
Sbjct: 187 LFDYIQGKNNYKQKIEMTAPVITEVSPSDGPFC-------------KSSFVVSFFVPKLN 327
Query: 207 GANLPLPKDSSVRIKEIPRKIVAVVSFSGFVND 239
AN P K + ++ VA F G VND
Sbjct: 328 QANPPPAK--GLHVQRWNNMYVAARQFGGHVND 420
>TC205430 similar to UP|O22600 (O22600) Glycine-rich protein, partial (19%)
Length = 421
Score = 30.0 bits (66), Expect = 1.3
Identities = 11/23 (47%), Positives = 17/23 (73%)
Frame = +3
Query: 269 VAQYNPPFTLPFQRRNEIALEVE 291
+ +YNPP+T+P R NE+ + VE
Sbjct: 180 LGRYNPPWTIPAFRTNEVMIPVE 248
>TC216256 homologue to UP|Q6PQQ3 (Q6PQQ3) AP2/EREBP transcription factor,
partial (35%)
Length = 1475
Score = 29.6 bits (65), Expect = 1.7
Identities = 23/112 (20%), Positives = 52/112 (45%), Gaps = 2/112 (1%)
Frame = +3
Query: 89 TLEEALMAVPDLETVKFKVLSRRDNYEIREVEPYFVAEATMP--GKSGFDFSGASQSFNV 146
T EEA A D+ +KF+ L+ N+++ + +A +T+P G SG + ++ S +
Sbjct: 840 TQEEAAEAY-DIAAIKFRGLNAVTNFDMSRYDVKSIANSTLPIGGLSGKN-KNSTDSASE 1013
Query: 147 LAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTM 198
+ ++ ++ ++ + S+ Q + P+ K + D W++
Sbjct: 1014SKSHEASRSDERDPSAASSVTFASQQQPSSSTLSFAIPI---KQDPSDYWSI 1160
>TC206112 similar to UP|Q8VIL5 (Q8VIL5) Ski proto-oncogene, partial (3%)
Length = 666
Score = 28.5 bits (62), Expect = 3.8
Identities = 22/55 (40%), Positives = 28/55 (50%)
Frame = +1
Query: 11 SQSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQANSLT 65
S SS T PN+ ++ SAS R RRRT S+ SSL A +S +S T
Sbjct: 115 SPSSSPTKTTMLPNLTVSVSASPRRR--LRRRTSSSTSFLSSLSSASSSSTSSPT 273
>TC223068
Length = 425
Score = 28.1 bits (61), Expect = 4.9
Identities = 22/72 (30%), Positives = 37/72 (50%), Gaps = 10/72 (13%)
Frame = +3
Query: 13 SSIRTSIPGRPNI--------AITNSASSSERISNRRRTMSA--VEARSSLILALASQAN 62
S + T+IPG A+++ A+ + + + R A +A+S L + L Q
Sbjct: 129 SGLPTNIPGSNTKTVIENELKAVSSVATQNTELPSARAWKPAPGFKAKSLLEIQLEEQKK 308
Query: 63 SLTQRLVVDVAT 74
SLT++LV +VAT
Sbjct: 309 SLTEKLVSEVAT 344
>TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein,
partial (58%)
Length = 935
Score = 28.1 bits (61), Expect = 4.9
Identities = 12/39 (30%), Positives = 19/39 (47%)
Frame = -3
Query: 252 ALKSDGQFKIKKGTSVEVAQYNPPFTLPFQRRNEIALEV 290
AL G +I++ + Y PP PFQ+ N I + +
Sbjct: 231 ALHQSGGTEIREANPSDHLPYTPPERFPFQKNNRITISL 115
>TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {Arabidopsis
thaliana;} , partial (42%)
Length = 1533
Score = 27.7 bits (60), Expect = 6.4
Identities = 15/44 (34%), Positives = 21/44 (47%)
Frame = -1
Query: 183 TPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRK 226
TP T + Q S +LP ++ + LPL K S K +P K
Sbjct: 777 TPTATLYLSCQPNALQSTLLP*RFSSRLPLDKKSYTSSKWLPSK 646
>BE330049 homologue to GP|2252828|gb| A_IG005I10.5 gene product {Arabidopsis
thaliana}, partial (54%)
Length = 467
Score = 27.3 bits (59), Expect = 8.4
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Frame = +1
Query: 6 NASLSSQSSIRTS---IPGRPNIAITNSASSSERISNRRRTMSAVEARSSL 53
+A+ SS S R+S PN++ + S+SSS ++R T ++ A SSL
Sbjct: 211 SAASSSSSPFRSSSSPTSSSPNLSASRSSSSSPSPASRSATSNSSHAPSSL 363
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.129 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,274,998
Number of Sequences: 63676
Number of extensions: 95440
Number of successful extensions: 455
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 447
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 449
length of query: 295
length of database: 12,639,632
effective HSP length: 97
effective length of query: 198
effective length of database: 6,463,060
effective search space: 1279685880
effective search space used: 1279685880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.19


