

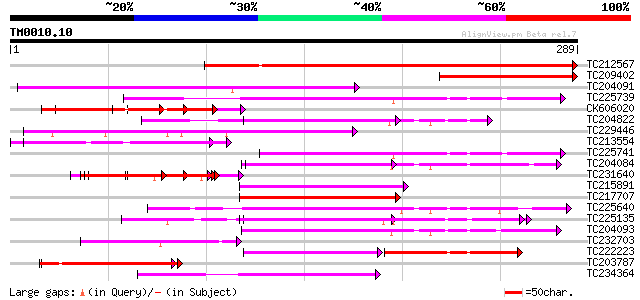
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.10
(289 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC212567 weakly similar to UP|PAB3_HUMAN (Q9H361) Polyadenylate-... 278 3e-75
TC209402 128 3e-30
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 82 4e-16
TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 70 9e-13
CK606020 66 2e-11
TC204822 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, part... 64 8e-11
TC229446 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 64 8e-11
TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protei... 63 2e-10
TC225741 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 60 1e-09
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 56 2e-08
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 56 2e-08
TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/ma... 56 2e-08
TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precu... 54 6e-08
TC225640 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, part... 54 6e-08
TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, ... 54 1e-07
TC204093 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprot... 54 1e-07
TC232703 similar to UP|Q93ZJ9 (Q93ZJ9) At1g80000/F19K16_3, parti... 54 1e-07
TC222223 similar to UP|Q8LKG6 (Q8LKG6) Cleavage stimulation fact... 53 2e-07
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 52 3e-07
TC234364 similar to UP|ROC5_NICSY (P19684) 33 kDa ribonucleoprot... 49 3e-06
>TC212567 weakly similar to UP|PAB3_HUMAN (Q9H361) Polyadenylate-binding
protein 3 (Poly(A)-binding protein 3) (PABP 3)
(Testis-specific poly(A)-binding protein), partial (5%)
Length = 615
Score = 278 bits (710), Expect = 3e-75
Identities = 131/190 (68%), Positives = 159/190 (82%)
Frame = +3
Query: 100 DEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGK 159
DE EK+KHAELL+LPPHGSEVYIGGI + S+EDL+ C+ +GEV+EVRIMK K E K
Sbjct: 39 DEDEKRKHAELLSLPPHGSEVYIGGIPH-ASDEDLKSLCERIGEVAEVRIMKGKXFFENK 215
Query: 160 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVA 219
G+ FV F + ELAS+AIEELNN+EF GKKIKCS SQAKHRLFIGN+P+ W VED+KK+V
Sbjct: 216 GFGFVTFTSVELASKAIEELNNTEFMGKKIKCSKSQAKHRLFIGNVPRSWGVEDLKKIVT 395
Query: 220 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAD 279
++GPGV +EL+KD++++ NRGFAFI+YYNHACAEYSRQKM + FKL NAPTVSWAD
Sbjct: 396 EIGPGVTGVELVKDMKNTNNNRGFAFIDYYNHACAEYSRQKMMSPTFKLGENAPTVSWAD 575
Query: 280 PRNSESSAVS 289
P+N+ESSA S
Sbjct: 576 PKNAESSAAS 605
>TC209402
Length = 1163
Score = 128 bits (321), Expect = 3e-30
Identities = 59/70 (84%), Positives = 66/70 (94%)
Frame = +1
Query: 220 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAD 279
++GPGVI +ELLKD Q+S RNRG+AFIEYYNHACAEYSRQKMSNSNFKL +NAPTVSWAD
Sbjct: 1 EIGPGVICVELLKDPQNSSRNRGYAFIEYYNHACAEYSRQKMSNSNFKLGSNAPTVSWAD 180
Query: 280 PRNSESSAVS 289
PRNSESSA+S
Sbjct: 181 PRNSESSAIS 210
Score = 33.1 bits (74), Expect = 0.15
Identities = 28/149 (18%), Positives = 64/149 (42%), Gaps = 13/149 (8%)
Frame = +1
Query: 153 KESGEGKGYAFVAFKTKELASQAIEELNNSEFK------------GKKIKCSSSQAKHRL 200
+ S +GYAF+ + A + ++++NS FK + + S+ +
Sbjct: 46 QNSSRNRGYAFIEYYNHACAEYSRQKMSNSNFKLGSNAPTVSWADPRNSESSAISLVKSV 225
Query: 201 FIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQK 260
++ N+P+ T + +K++ G I+ +L +S F F+ + + A +
Sbjct: 226 YVKNLPENITQDRLKELFEH--HGKITKVVLPSAKSGQEKSRFGFVHFAERSSA--MKAL 393
Query: 261 MSNSNFKLENNAPTVSWADPR-NSESSAV 288
+ ++++ S A P+ NS+ A+
Sbjct: 394 KNTEKYEIDGQLLECSLAKPQANSQKPAL 480
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 81.6 bits (200), Expect = 4e-16
Identities = 54/193 (27%), Positives = 96/193 (48%), Gaps = 19/193 (9%)
Frame = +2
Query: 5 PGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEV 64
P R+R V ++ S E + ++P + +++ ++ EE E EE EEEE E+EE
Sbjct: 26 PMGRKRKVAPKSSLSTEPPLKQQQPKPENPPQEYEEVEEEVEEEEVEEEVEEEEEEEEED 205
Query: 65 EEVEEEEEEEEEEEE-VEEESKPSDGEEEMRVDHTNDEVEKKKHAELLA----------- 112
EE EEEEEEEEEE + V+++ + E+++ + + + K + LL
Sbjct: 206 EEEEEEEEEEEEENDVVKQQQQQQQDEDDIPISKLLEPLGKDQLLNLLCDAAAKHRDVEE 385
Query: 113 -------LPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVA 165
P ++++ G+ + + L + GE+ + + + K SG+ KGY F+
Sbjct: 386 RIRKAADGDPVHRKIFVHGLGWDTTAGTLISAFRQYGEIEDCKAVTDKVSGKSKGYGFIL 565
Query: 166 FKTKELASQAIEE 178
FKT+ A A++E
Sbjct: 566 FKTRRGAQNALKE 604
Score = 38.9 bits (89), Expect = 0.003
Identities = 18/76 (23%), Positives = 39/76 (50%)
Frame = +2
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++Y+ + ++ + L F GE+ E + K +G+ KG+ ++ E A +A+EE
Sbjct: 737 KIYVSNVGADLDPQKLLAFFSRFGEIEEGPLGLDKATGKPKGFCLFVYRNPESARRALEE 916
Query: 179 LNNSEFKGKKIKCSSS 194
+ +F+G + C +
Sbjct: 917 -PHKDFEGHILHCQKA 961
>TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (85%)
Length = 1436
Score = 70.5 bits (171), Expect = 9e-13
Identities = 57/236 (24%), Positives = 101/236 (42%), Gaps = 11/236 (4%)
Frame = +3
Query: 59 VEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGS 118
V+D+ VEEE+ EE E E V E+ S +
Sbjct: 405 VDDDGAGLVEEEKVEENEGEAVAEQDSDSSA----------------------------T 500
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++Y G + +V L Q G + ++ +++G+ +G+AFV E + IE
Sbjct: 501 KLYFGNLPYSVDSAKLAGLIQDYGSAELIEVLYDRDTGKSRGFAFVTMSCIEDCNAVIEN 680
Query: 179 LNNSEFKGKKIKCSSS-----------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVIS 227
L+ EF G+ ++ + S + +H+LF+GN+ T E + + + G V+
Sbjct: 681 LDGKEFLGRTLRVNFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGT-VVG 857
Query: 228 IELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNS 283
+L D +GR+RG+ F+ Y AE + ++ +LE A VS A + +
Sbjct: 858 ARVLYD-GETGRSRGYGFVCYSTK--AEMEAALAALNDVELEGRAMRVSLAQGKRA 1016
>CK606020
Length = 340
Score = 66.2 bits (160), Expect = 2e-11
Identities = 38/68 (55%), Positives = 47/68 (68%)
Frame = +1
Query: 24 SEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEE 83
S P+ P +D+++D D+EEE E EE EEEEE E+EE+EE EEEEEEE EEE EEE
Sbjct: 145 SLPDSPVANDDEID--PSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEEE 318
Query: 84 SKPSDGEE 91
+GEE
Sbjct: 319 ---VEGEE 333
Score = 62.0 bits (149), Expect = 3e-10
Identities = 31/63 (49%), Positives = 43/63 (68%)
Frame = +1
Query: 17 PESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEE 76
P+SP N + P+ +E+ + + + ++EEE E EE+EEEEE E+EEVEE EEEE E E
Sbjct: 151 PDSPVANDDEIDPSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEEEVEGE 330
Query: 77 EEE 79
E E
Sbjct: 331 EVE 339
Score = 45.8 bits (107), Expect = 2e-05
Identities = 25/54 (46%), Positives = 37/54 (68%)
Frame = +1
Query: 53 VEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKK 106
V ++E+ D ++E EEEEEEEEEEEE EEE + + EEE + +EVE+++
Sbjct: 163 VANDDEI-DPSLDEEEEEEEEEEEEEE-EEEEEEIEEEEEEEEEEVEEEVEEEE 318
Score = 42.4 bits (98), Expect = 2e-04
Identities = 24/60 (40%), Positives = 34/60 (56%)
Frame = +1
Query: 61 DEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEV 120
D+E++ +EEEEEEEEEE EEE EEE ++ +E E++ E+ G EV
Sbjct: 172 DDEIDPSLDEEEEEEEEEEEEEEE-----EEEEEIEEEEEEEEEEVEEEVEEEEVEGEEV 336
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/61 (36%), Positives = 33/61 (54%)
Frame = +1
Query: 5 PGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEV 64
P ++ E E E E+ + +E+ + + + ++EEE EEV EEEEVE EEV
Sbjct: 160 PVANDDEIDPSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEV-EEEEVEGEEV 336
Query: 65 E 65
E
Sbjct: 337 E 339
>TC204822 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (67%)
Length = 1232
Score = 63.9 bits (154), Expect = 8e-11
Identities = 45/197 (22%), Positives = 92/197 (45%), Gaps = 18/197 (9%)
Frame = +3
Query: 68 EEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIAN 127
++EEE+ E E ES +GE+ E PP +++++G +
Sbjct: 222 QQEEEQSLAETEAGLESWEPNGEDA--------------GDESFVEPPEEAKLFVGNLPY 359
Query: 128 NVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGK 187
+V + L + + G V ++ +E+ + +G+ FV T E A A+E+ N + G+
Sbjct: 360 DVDSQKLAMLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSTVEEAESAVEKFNRYDIDGR 539
Query: 188 KIKCS----------------SSQAKHRLFIGNIPKKWTVED--MKKVVADVGPGVISIE 229
+ + S ++ +++GN+P W V++ +K++ + G V++
Sbjct: 540 LLTVNKASPRGTRPERPPPRRSFESSLSIYVGNLP--WDVDNTRLKQIFSKHG-NVVNAR 710
Query: 230 LLKDLQSSGRNRGFAFI 246
++ D + SGR+RGF F+
Sbjct: 711 VVYD-RESGRSRGFGFV 758
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/80 (25%), Positives = 38/80 (47%)
Frame = +3
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+Y+G + +V L+ G V R++ +ESG +G+ FV + + A+ L
Sbjct: 624 IYVGNLPWDVDNTRLKQIFSKHGNVVNARVVYDRESGRSRGFGFVTMSDETEMNDAVAAL 803
Query: 180 NNSEFKGKKIKCSSSQAKHR 199
+ G+ IK S ++ + R
Sbjct: 804 DGESLDGRAIKVSVAEDRPR 863
>TC229446 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(20%)
Length = 850
Score = 63.9 bits (154), Expect = 8e-11
Identities = 54/227 (23%), Positives = 96/227 (41%), Gaps = 57/227 (25%)
Frame = +3
Query: 8 RQRDVTAQNPESP---EGNSEPEKPTDSDEQVDFDGDNDQEEE---IEYEEVEEEEEVED 61
++R + + +PE E + ++P + D+Q E+ + +E ++EEE ED
Sbjct: 90 KKRKLRSSDPEPTKPIEPQQQDQEPVELDQQQQQPNPEQTLEDPNTMAVDEPKQEEEEED 269
Query: 62 EEVEEVEEEEEEEEEEEE--------VEEESKP----------------SDGEEEMRVDH 97
EE + E EEEEEE EE+ +EE+ P + EEE D
Sbjct: 270 EEPQNAESEEEEEEAEEQQEERPPETLEEQQDPETLAAANHAEVNGGNEGNNEEEEEEDL 449
Query: 98 TNDEVEKKKHAE---------------------------LLALPPHGSEVYIGGIANNVS 130
T +E +K E L + P ++++ G+ + +
Sbjct: 450 TLEEEPVEKLLEPFTKEQLHSLVTQAVDMFPEFVDSVRRLADVDPAHRKIFVHGLGWDAT 629
Query: 131 EEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
+ L GE+ + + + K SG+ KGYAF+ FK ++ A +A++
Sbjct: 630 ADTLTAVFGKYGEIEDCKAVTDKVSGKSKGYAFILFKHRDDARKALK 770
>TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1,
partial (18%)
Length = 451
Score = 62.8 bits (151), Expect = 2e-10
Identities = 37/104 (35%), Positives = 57/104 (54%)
Frame = +2
Query: 1 MHKMPGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVE 60
+H P + + + +P+ + E P + + + EEE+E EEVEEEE
Sbjct: 59 LHSTPINQILAMGRKRKVAPKSSVSAEPPLKQHQPKPENAAQEYEEEVE-EEVEEEEV-- 229
Query: 61 DEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEK 104
+EEVEE EEEEE+EE+EEE EE+ K ++ + D +D + K
Sbjct: 230 EEEVEEEEEEEEDEEDEEEEEEDQKDQYQHQQQQQDEDDDPISK 361
Score = 53.5 bits (127), Expect = 1e-07
Identities = 35/106 (33%), Positives = 59/106 (55%)
Frame = +2
Query: 8 RQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEV 67
R+R V ++ S E + +P + +++ + EEE+E EEVEEE E EE EE
Sbjct: 98 RKRKVAPKSSVSAEPPLKQHQPKPENAAQEYE--EEVEEEVEEEEVEEEVE---EEEEEE 262
Query: 68 EEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLAL 113
E+EE+EEEEEE+ +++ + +++ D + +E +LL L
Sbjct: 263 EDEEDEEEEEEDQKDQYQHQQQQQDEDDDPISKLLEPLSKDQLLIL 400
>TC225741 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (57%)
Length = 857
Score = 60.1 bits (144), Expect = 1e-09
Identities = 43/167 (25%), Positives = 78/167 (45%), Gaps = 11/167 (6%)
Frame = +2
Query: 128 NVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGK 187
+V L Q G + ++ ++SG+ +G+AFV E + IE L+ EF G+
Sbjct: 2 SVDSXKLAGLIQDYGSAELIEVLYDRDSGKSRGFAFVTMSCIEDCNAVIENLDGKEFLGR 181
Query: 188 KIKCSSS-----------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQS 236
++ + S + +H+LF+GN+ T E + + + G V+ +L D
Sbjct: 182 XLRVNFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGT-VVGARVLYD-GE 355
Query: 237 SGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNS 283
+GR+RG+ F+ Y AE + ++ +LE A VS A + +
Sbjct: 356 TGRSRGYGFVCYSTQ--AEMEAAVAALNDVELEGRAMRVSLAQGKRA 490
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 56.2 bits (134), Expect = 2e-08
Identities = 43/191 (22%), Positives = 86/191 (44%), Gaps = 28/191 (14%)
Frame = -1
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++++G + +V L +S G V V ++ K +G +G+ FV + E A A ++
Sbjct: 878 KLFVGNLPFSVDSARLAELFESAGNVEVVEVIYDKTTGRSRGFGFVTMSSVEEAEAAAKQ 699
Query: 179 LNNSEFKGKKIKCSS--------------------------SQAKHRLFIGNIPKKWTVE 212
N E G+ ++ +S S +++R+ +GN+ W V+
Sbjct: 698 FNGYELDGRSLRVNSGPPPARNESAPRFRGGSSFGSRGGGPSDSENRVHVGNLA--WGVD 525
Query: 213 D--MKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLEN 270
D ++ + + G V+ ++ D + SGR+RGF F+ + + + + Q + + L
Sbjct: 524 DVALESLFREQGKKVLEARVIYD-RESGRSRGFGFVTFGSPDEVKSAIQSLDGVD--LNG 354
Query: 271 NAPTVSWADPR 281
A VS AD +
Sbjct: 353 RAIRVSLADSK 321
Score = 46.6 bits (109), Expect = 1e-05
Identities = 20/77 (25%), Positives = 40/77 (50%)
Frame = +2
Query: 121 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELN 180
++GG+A + L GE+ E +I+ +E+G +G+ FV F +++ AI +N
Sbjct: 1250 FVGGLAWATDDHALERAFSQYGEIVETKIINDRETGRSRGFGFVTFASEQSMKDAIGAMN 1429
Query: 181 NSEFKGKKIKCSSSQAK 197
G+ I + +Q++
Sbjct: 1430 GQNLDGRNITVNEAQSR 1480
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 55.8 bits (133), Expect = 2e-08
Identities = 34/68 (50%), Positives = 43/68 (63%), Gaps = 3/68 (4%)
Frame = +1
Query: 41 DNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVD---H 97
DN + E EE +EEEE E+EE EE EEEEEEEEE+EE EEE +G + +D H
Sbjct: 1 DNVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEE----EGNVLVIIDEDKH 168
Query: 98 TNDEVEKK 105
+E+ KK
Sbjct: 169 DTEELNKK 192
Score = 51.2 bits (121), Expect = 5e-07
Identities = 26/42 (61%), Positives = 31/42 (72%)
Frame = +1
Query: 39 DGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEV 80
D ++EE+ E EE EEEEE E+EE EE EE+EEEEEEE V
Sbjct: 16 DTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNV 141
Score = 48.1 bits (113), Expect = 5e-06
Identities = 27/55 (49%), Positives = 34/55 (61%)
Frame = +1
Query: 37 DFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEE 91
D + +++EE E EE EEEEE E+EE EE +EEEEEEE V + D EE
Sbjct: 16 DTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEE 180
Score = 47.8 bits (112), Expect = 6e-06
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 6/78 (7%)
Frame = +1
Query: 32 SDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEE------EEEEEEEVEEESK 85
SD Q + + ++EEE E EE EEEEE E+EE EE EEEE +E++ + E K
Sbjct: 13 SDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEELNKK 192
Query: 86 PSDGEEEMRVDHTNDEVE 103
D + M+ +++ +E
Sbjct: 193 CEDFIKRMKATFSSNNLE 246
Score = 46.6 bits (109), Expect = 1e-05
Identities = 24/48 (50%), Positives = 31/48 (64%)
Frame = +1
Query: 60 EDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKH 107
E+EE +E EEEEEEEEEEEE EEE + EEE + +++ KH
Sbjct: 25 EEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKH 168
Score = 42.0 bits (97), Expect = 3e-04
Identities = 21/59 (35%), Positives = 33/59 (55%)
Frame = +1
Query: 61 DEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSE 119
D + + +EEEEE+EEEEE EEE + + EEE + +E E+ ++ H +E
Sbjct: 1 DNVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTE 177
Score = 39.3 bits (90), Expect = 0.002
Identities = 18/52 (34%), Positives = 33/52 (62%)
Frame = +1
Query: 25 EPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEE 76
E E+ + +E+ + + + ++EEE E EE EEEEE E + ++E++ + EE
Sbjct: 25 EEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEE 180
Score = 37.4 bits (85), Expect = 0.008
Identities = 17/55 (30%), Positives = 35/55 (62%)
Frame = +1
Query: 23 NSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEE 77
+++ E+ +E+ + + + ++EEE E EE +EEEE E+ V + +E++ + EE
Sbjct: 16 DTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEE 180
Score = 35.4 bits (80), Expect = 0.030
Identities = 21/66 (31%), Positives = 36/66 (53%), Gaps = 5/66 (7%)
Frame = +1
Query: 16 NPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEE-----VEDEEVEEVEEE 70
N S E EK + +E+ + + + ++EEE E +E EEEEE + DE+ + EE
Sbjct: 4 NVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEEL 183
Query: 71 EEEEEE 76
++ E+
Sbjct: 184NKKCED 201
Score = 34.3 bits (77), Expect = 0.068
Identities = 18/58 (31%), Positives = 35/58 (60%)
Frame = +1
Query: 25 EPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEE 82
E E+ + +E+ + + + ++EEE E E+ EEEEE + V E++ + EE ++ E+
Sbjct: 28 EEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEELNKKCED 201
>TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/maf19_30
{Arabidopsis thaliana;} , partial (41%)
Length = 782
Score = 55.8 bits (133), Expect = 2e-08
Identities = 25/86 (29%), Positives = 51/86 (59%)
Frame = +2
Query: 118 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
++++IGG++ + E+ LR GEV + RI+ +E+G +G+ F+ + + E AS AI+
Sbjct: 173 TKLFIGGVSYSTDEQSLREAFSKYGEVVDARIIMDRETGRSRGFGFITYTSVEEASSAIQ 352
Query: 178 ELNNSEFKGKKIKCSSSQAKHRLFIG 203
L+ + G+ I+ + + + R + G
Sbjct: 353 ALDGQDLHGRPIRVNYANERPRGYGG 430
>TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precursor,
partial (35%)
Length = 1008
Score = 54.3 bits (129), Expect = 6e-08
Identities = 23/82 (28%), Positives = 50/82 (60%)
Frame = +1
Query: 118 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
+++++GGI+ + + LR GEV +V+++ +E+G +G+ F+ F T E AS AI+
Sbjct: 163 AKLFVGGISYSTDDMSLRESPARYGEVIDVKVIMDRETGRSRGFGFITFATSEDASYAIQ 342
Query: 178 ELNNSEFKGKKIKCSSSQAKHR 199
++ + G++I+ + + + R
Sbjct: 343 GMDGQDLHGRRIRVNYATERSR 408
>TC225640 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (66%)
Length = 1290
Score = 54.3 bits (129), Expect = 6e-08
Identities = 55/240 (22%), Positives = 100/240 (40%), Gaps = 24/240 (10%)
Frame = +3
Query: 71 EEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVS 130
+EEEEEE E + + G EE H +K + ++++G + ++
Sbjct: 333 KEEEEEETSWENQGDTAWGTEE--GGHEEGGFAEKAEED---------KIFVGNLPFDID 479
Query: 131 EEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIK 190
E+L G V ++ + + +G+ FV T E +A+E + E G+ +
Sbjct: 480 SENLASLFGQAGTVEVAEVIYNRATDRSRGFGFVTMSTLEELKKAVEMFSGYELNGRVLT 659
Query: 191 CSSSQAKH-----------------RLFIGNIPKKWTVED--MKKVVADVGPGVISIELL 231
+ + K R+++GN+P W V+D ++++ ++ G V ++
Sbjct: 660 VNKAAPKGAQPERPPRPPRSFSSGLRVYVGNLP--WEVDDARLEQIFSEHGK-VEDARVV 830
Query: 232 KDLQSSGRNRGFAFIEY-----YNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNSESS 286
D + +GR+RGF F+ N A A Q L+ A V+ A R S SS
Sbjct: 831 YD-RETGRSRGFGFVTMSSETDMNDAIAALDGQ-------SLDGRAIRVNVAQDRPSRSS 986
>TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(97%)
Length = 2392
Score = 53.5 bits (127), Expect = 1e-07
Identities = 34/153 (22%), Positives = 76/153 (49%), Gaps = 8/153 (5%)
Frame = +3
Query: 118 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
+ +Y+G + NV++ L +G+V VR+ + S GY +V F + A++A++
Sbjct: 180 TSLYVGDLDPNVTDAQLYDLFNQLGQVVSVRVCRDLTSRRSLGYGYVNFSNPQDAARALD 359
Query: 178 ELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIE 229
LN + + I+ S + +FI N+ + + + + G ++S +
Sbjct: 360 VLNFTPLNNRPIRIMYSHRDPSIRKSGQGNIFIKNLDRAIDHKALHDTFSTFG-NILSCK 536
Query: 230 LLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMS 262
+ D SSG+++G+ F+++ N A+ + +K++
Sbjct: 537 VATD--SSGQSKGYGFVQFDNEESAQKAIEKLN 629
Score = 48.9 bits (115), Expect = 3e-06
Identities = 34/159 (21%), Positives = 75/159 (46%), Gaps = 12/159 (7%)
Frame = +3
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
++I + + + L + G + ++ SG+ KGY FV F +E A +AIE+L
Sbjct: 450 IFIKNLDRAIDHKALHDTFSTFGNILSCKVA-TDSSGQSKGYGFVQFDNEESAQKAIEKL 626
Query: 180 NNSEFKGKKI-----------KCSSSQAK-HRLFIGNIPKKWTVEDMKKVVADVGPGVIS 227
N K++ + ++ +AK + +F+ N+ + T +++K + G + S
Sbjct: 627 NGMLLNDKQVYVGPFLRKQERESAADKAKFNNVFVKNLSESTTDDELKNTFGEFGT-ITS 803
Query: 228 IELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNF 266
+++D G+++ F F+ + N A + + ++ F
Sbjct: 804 AVVMRD--GDGKSKCFGFVNFENADDAARAVEALNGKKF 914
Score = 45.8 bits (107), Expect = 2e-05
Identities = 35/142 (24%), Positives = 69/142 (47%), Gaps = 2/142 (1%)
Frame = +3
Query: 58 EVEDEEVEEVEEEEEEEEEEEE--VEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPP 115
E D+ VE ++ +++E V + K S+ E E++ E K+ A+
Sbjct: 861 ENADDAARAVEALNGKKFDDKEWYVGKAQKKSERENELK---QRFEQSMKEAADKY---- 1019
Query: 116 HGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQA 175
G+ +Y+ + +++ +E L+ G ++ ++M+ +G +G FVAF T E AS+A
Sbjct: 1020QGANLYVKNLDDSIGDEKLKELFSPFGTITSCKVMR-DPNGLSRGSGFVAFSTPEEASRA 1196
Query: 176 IEELNNSEFKGKKIKCSSSQAK 197
+ E+N K + + +Q K
Sbjct: 1197LLEMNGKMVVSKPLYVTLAQRK 1262
>TC204093 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A,
chloroplast precursor (CP29A), partial (63%)
Length = 1403
Score = 53.5 bits (127), Expect = 1e-07
Identities = 44/191 (23%), Positives = 84/191 (43%), Gaps = 28/191 (14%)
Frame = +2
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++++G + NV L +S G V V ++ K +G +G+ FV + E A A ++
Sbjct: 332 KLFVGNLPFNVDSAQLAELFESAGNVEVVEVIYDKTTGRSRGFGFVTMSSVEEAEAAAQQ 511
Query: 179 LNNSEFKGKKIKCSS--------------------------SQAKHRLFIGNIPKKWTVE 212
N E G+ ++ +S S +++R+ + N+ W V+
Sbjct: 512 FNGYELDGRALRVNSGPPPARNESAPRFRGGSSFGSRGGGPSDSENRVHVSNLA--WGVD 685
Query: 213 D--MKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLEN 270
+ +K + + G V+ ++ D + SGR+RGF F+ + + E + S + L
Sbjct: 686 NVALKSLFREQG-NVLEARVIYD-RESGRSRGFGFVTF--SSPDEVNSAIQSLNGVDLNG 853
Query: 271 NAPTVSWADPR 281
A VS AD +
Sbjct: 854 RAIRVSLADSK 886
>TC232703 similar to UP|Q93ZJ9 (Q93ZJ9) At1g80000/F19K16_3, partial (10%)
Length = 527
Score = 53.5 bits (127), Expect = 1e-07
Identities = 34/84 (40%), Positives = 48/84 (56%), Gaps = 2/84 (2%)
Frame = +1
Query: 37 DFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE--EEEEVEEESKPSDGEEEMR 94
D+D + D EEE E E EEEEEVE+E++EE E E E E V ++S +D + +
Sbjct: 4 DYDDEEDLEEEEEAVEEEEEEEVEEEDLEEEERSEHEYEGGANGTVVKDSDDADVKALLE 183
Query: 95 VDHTNDEVEKKKHAELLALPPHGS 118
DE EKK++ E A+P G+
Sbjct: 184 ESGNGDEEEKKEN-EPFAVPTAGA 252
>TC222223 similar to UP|Q8LKG6 (Q8LKG6) Cleavage stimulation factor 64,
partial (22%)
Length = 445
Score = 52.8 bits (125), Expect = 2e-07
Identities = 25/71 (35%), Positives = 40/71 (56%)
Frame = +2
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
V++G I + +EE L CQ VG V R++ +E+G+ KGY F +K +E A A L
Sbjct: 164 VFVGNIPYDATEEQLIEICQEVGPVVSFRLVIDRETGKPKGYGFCEYKDEETALSARRNL 343
Query: 180 NNSEFKGKKIK 190
E G++++
Sbjct: 344 QGYEINGRQLR 376
Score = 45.1 bits (105), Expect = 4e-05
Identities = 25/71 (35%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Frame = +2
Query: 192 SSSQAKHR-LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYN 250
++SQ++HR +F+GNIP T E + ++ +VGP V+S L+ D + +G+ +G+ F EY +
Sbjct: 137 ATSQSQHRCVFVGNIPYDATEEQLIEICQEVGP-VVSFRLVID-RETGKPKGYGFCEYKD 310
Query: 251 HACAEYSRQKM 261
A +R+ +
Sbjct: 311 EETALSARRNL 343
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 52.0 bits (123), Expect = 3e-07
Identities = 28/69 (40%), Positives = 40/69 (57%)
Frame = -3
Query: 17 PESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEE 76
PE G +E D DE D D D+D E++ + +E +E E EDE+ E EE EE+EE+
Sbjct: 487 PEGGAGGAEENGEEDEDEDGD-DQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEED 311
Query: 77 EEEVEEESK 85
EE ++ K
Sbjct: 310 EEALQPPKK 284
Score = 40.8 bits (94), Expect = 7e-04
Identities = 19/73 (26%), Positives = 44/73 (60%)
Frame = -3
Query: 16 NPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE 75
N + + NS+ + P + +G+ D++E+ + ++ ++E++ +D+E +E EEE+E+
Sbjct: 523 NNSNNKSNSK-KAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDG 347
Query: 76 EEEEVEEESKPSD 88
E+E EE + +
Sbjct: 346 AEDEENEEDEEDE 308
Score = 37.4 bits (85), Expect = 0.008
Identities = 26/117 (22%), Positives = 53/117 (45%), Gaps = 26/117 (22%)
Frame = -3
Query: 18 ESPEGNSEPEKPTDSDEQVDFDGDN---DQEEEIEYEEV--------------------- 53
E + ++E E D + D DGD + EEE+ E+
Sbjct: 652 EDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGA 473
Query: 54 --EEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHA 108
EE EDE+ + ++++++E+++++ EE+ + +E+ D N+E E+ + A
Sbjct: 472 GGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEEA 302
Score = 35.8 bits (81), Expect = 0.023
Identities = 25/127 (19%), Positives = 56/127 (43%), Gaps = 29/127 (22%)
Frame = -3
Query: 18 ESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVED---------------- 61
ES + + ++ + ++ D +GD+D + + + E EEE ED
Sbjct: 670 ESKIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKK 491
Query: 62 -------------EEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHA 108
EE E+ + +++++++E++ +++ + GEEE ++E E+ +
Sbjct: 490 APEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEED 311
Query: 109 ELLALPP 115
E PP
Sbjct: 310 EEALQPP 290
Score = 35.0 bits (79), Expect = 0.040
Identities = 21/90 (23%), Positives = 48/90 (53%), Gaps = 3/90 (3%)
Frame = -3
Query: 6 GTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVE 65
G + ++++++ NS + + + G + EE E E+ +++++ D+E +
Sbjct: 571 GEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDD--DDEDD 398
Query: 66 EVEEEEEE---EEEEEEVEEESKPSDGEEE 92
+ ++EE+E EE+E+ E+E D E+E
Sbjct: 397 DDDDEEDEAGEEEDEDGAEDEENEEDEEDE 308
>TC234364 similar to UP|ROC5_NICSY (P19684) 33 kDa ribonucleoprotein,
chloroplast precursor, partial (32%)
Length = 710
Score = 48.5 bits (114), Expect = 3e-06
Identities = 28/124 (22%), Positives = 57/124 (45%)
Frame = +2
Query: 66 EVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGI 125
EV ++ E +++E E + ++ EEE +V +ND +Y+G +
Sbjct: 269 EVAQDTTESQQDEPEPETVEKTEQEEEQKVSDSND----------------AGRLYVGNL 400
Query: 126 ANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFK 185
+++ +L G V+ V I+ + + +G+AFV + E A +AI + S+
Sbjct: 401 PYSITNSELGELFGEAGTVASVEIVYDRVTDRSRGFAFVTMGSVEDAKEAIRMFDGSQVG 580
Query: 186 GKKI 189
G+ +
Sbjct: 581 GRTV 592
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.303 0.125 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,248,300
Number of Sequences: 63676
Number of extensions: 129832
Number of successful extensions: 6968
Number of sequences better than 10.0: 672
Number of HSP's better than 10.0 without gapping: 1860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4148
length of query: 289
length of database: 12,639,632
effective HSP length: 96
effective length of query: 193
effective length of database: 6,526,736
effective search space: 1259660048
effective search space used: 1259660048
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.10


