

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
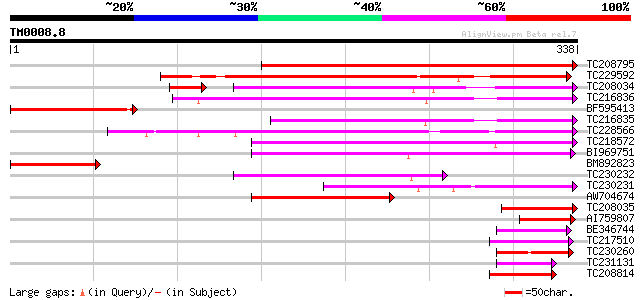
Query= TM0008.8
(338 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208795 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (28%) 315 2e-86
TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding pr... 172 1e-43
TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 144 2e-36
TC216836 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 149 2e-36
BF595413 similar to SP|Q40872|AG_PA Floral homeotic protein AGAM... 126 1e-29
TC216835 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 126 2e-29
TC228566 weakly similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thalia... 118 4e-27
TC218572 similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana geno... 114 6e-26
BI969751 107 1e-23
BM892823 similar to GP|22946353|gb CG6214-PA {Drosophila melanog... 94 7e-20
TC230232 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-li... 84 1e-16
TC230231 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-li... 80 1e-15
AW704674 66 3e-11
TC208035 similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (15%) 58 5e-09
AI759807 50 1e-06
BE346744 weakly similar to GP|21593126|gb| inhibitor of apoptosi... 47 1e-05
TC217510 44 8e-05
TC230260 44 1e-04
TC231131 similar to GB|BAB71852.1|16902294|AB062739 kinesin-rela... 43 2e-04
TC208814 43 2e-04
>TC208795 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (28%)
Length = 799
Score = 315 bits (806), Expect = 2e-86
Identities = 152/188 (80%), Positives = 167/188 (87%)
Frame = +2
Query: 151 HYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQ 210
+Y QKEQLS+GVRDMKQKHMA+LLTSIEKGI KL+EK+ EIENMNRKNRELAERIKQ
Sbjct: 17 NYFYTQKEQLSKGVRDMKQKHMAALLTSIEKGISTKLKEKDVEIENMNRKNRELAERIKQ 196
Query: 211 VAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLS 270
VAVE Q+WHYRAKYNES+VN LRNNLQQAIS G EQGKEGFGDSEVDD ASYIDPNNFL+
Sbjct: 197 VAVEVQSWHYRAKYNESIVNTLRNNLQQAISQGAEQGKEGFGDSEVDDDASYIDPNNFLN 376
Query: 271 IPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKT 330
I AP+ S H YQ MENL+CRACK K VSMLL+PCRHL LCKDC+GFINVCP+CQ+IKT
Sbjct: 377 ILAAPINSTHKSYQDMENLTCRACKVKTVSMLLMPCRHLCLCKDCEGFINVCPICQLIKT 556
Query: 331 ASVEVYLS 338
ASVEV+LS
Sbjct: 557 ASVEVHLS 580
>TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding protein SBP1
(Fragment), partial (91%)
Length = 1267
Score = 172 bits (437), Expect = 1e-43
Identities = 96/249 (38%), Positives = 151/249 (60%), Gaps = 4/249 (1%)
Frame = +1
Query: 91 SASIPNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELD 150
S P ++STGL LS D N+ +TS S +++ +GD+I+ EL +Q E+D
Sbjct: 358 SVDFLQPRSVSTGLGLSLD----NTHLTSTGDS-----ALLSLIGDDIKRELQQQDAEID 510
Query: 151 HYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQ 210
++K+Q E+L + V + Q ++ IE +++KLREKE +E++N++N EL ++++Q
Sbjct: 511 RFLKVQGERLRQAVLEKVQATQLQSVSLIEDKVLQKLREKEAMVESINKRNIELEDQMEQ 690
Query: 211 VAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPN---- 266
+ VEA +W RA+YNE+++ AL+ NLQQA KEG GDSEVDD AS +
Sbjct: 691 LTVEAGSWQQRARYNENMIAALKFNLQQAYVQS-RDSKEGCGDSEVDDTASCCNGRSLDF 867
Query: 267 NFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
+ LS MK E ++C+AC+ +V+M+L+PC+HL LCKDC+ ++ CP+CQ
Sbjct: 868 HLLSRENTDMK---------EMMTCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 1020
Query: 327 MIKTASVEV 335
K +EV
Sbjct: 1021SSKFIGMEV 1047
>TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1183
Score = 144 bits (363), Expect(2) = 2e-36
Identities = 75/210 (35%), Positives = 118/210 (55%), Gaps = 5/210 (2%)
Frame = +3
Query: 134 LGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEE 193
L + + +++ +Q++E+D +++ Q+EQL R + + +Q+H +LL + E+ ++++LREKE E
Sbjct: 318 LSEGLSSQIKQQRDEIDQFLQAQEEQLRRALAEKRQRHYRTLLRAAEESVLRRLREKEAE 497
Query: 194 IENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQA--ISHGVEQGKEGF 251
+E R N EL R Q++VEAQ W RAK E+ AL+ LQQA I G G G
Sbjct: 498 LEKATRHNAELEARATQLSVEAQLWQARAKAQEATAAALQAQLQQAMMIGDGENGGGGGL 677
Query: 252 ---GDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRH 308
G D ++Y+DP+ P CR C + S++++PCRH
Sbjct: 678 SCAGGGAEDAESAYVDPDRVGPTP-----------------KCRGCAKRVASVVVLPCRH 806
Query: 309 LSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
L +C +CD CPVC +K ++VEVYLS
Sbjct: 807 LCICAECDTHFRACPVCLTVKNSTVEVYLS 896
Score = 26.2 bits (56), Expect(2) = 2e-36
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = +2
Query: 96 NPNALSTGLRLSYDDDERNSSV 117
N N +STGLRLS+ D ++ +
Sbjct: 200 NQNVVSTGLRLSFGDQQQRQQL 265
>TC216836 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1272
Score = 149 bits (375), Expect = 2e-36
Identities = 89/254 (35%), Positives = 139/254 (54%), Gaps = 13/254 (5%)
Frame = +1
Query: 98 NALSTGLRLSYDDD-----ERNSSVTSASGSMTAPPSIILSL-GDNIRTELDRQQEELDH 151
N +STGLRLS+DD +R + S + S LSL + +++ +Q++E+D
Sbjct: 250 NVVSTGLRLSFDDQHFQQQQRLQLHQNESQQHRSHSSAFLSLLSQGLGSQIKQQRDEIDQ 429
Query: 152 YIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQV 211
+ Q EQL R + + +Q+H +LL++ E+ + ++LREKE E+E RKN EL R ++
Sbjct: 430 LLHAQGEQLRRALAEKRQRHYRALLSAAEEAVARQLREKEAEVEMATRKNAELEARAAKL 609
Query: 212 AVEAQNWHYRAKYNESVVNALRNNLQQAI-SHGVEQG-----KEGFGDSEVDDAAS-YID 264
+VEAQ W +A+ E+ +L+ LQQ I SHG E + + +DA S YID
Sbjct: 610 SVEAQVWQAKARAQEATAVSLQTKLQQTILSHGGEDPAVVGVSSAAVEGQAEDAESAYID 789
Query: 265 PNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPV 324
P+ ++ A K CR C + S++++PCRHL +C +CD CPV
Sbjct: 790 PDRVVAATAARPK-------------CRGCAKRVASVVVLPCRHLCVCTECDAHFRACPV 930
Query: 325 CQMIKTASVEVYLS 338
C K ++VEV+LS
Sbjct: 931 CLTPKNSTVEVFLS 972
>BF595413 similar to SP|Q40872|AG_PA Floral homeotic protein AGAMOUS (GAG2).
[Korean ginseng] {Panax ginseng}, partial (6%)
Length = 422
Score = 126 bits (317), Expect = 1e-29
Identities = 64/76 (84%), Positives = 68/76 (89%)
Frame = +2
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNATNQLQLFGNLQAGRNVDPVSYIGNEHISSMIQPN 60
M GGNNGN LLPVFLDE QFQ QTN +NQLQLFGNLQAG +VDPV+Y GNEHISSMIQPN
Sbjct: 197 MLGGNNGNCLLPVFLDETQFQHQTNVSNQLQLFGNLQAGCSVDPVNYFGNEHISSMIQPN 376
Query: 61 KRSREMEDISKKQRLQ 76
KRS+EMEDIS KQRLQ
Sbjct: 377 KRSKEMEDIS-KQRLQ 421
>TC216835 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (47%)
Length = 776
Score = 126 bits (316), Expect = 2e-29
Identities = 70/189 (37%), Positives = 105/189 (55%), Gaps = 6/189 (3%)
Frame = +3
Query: 156 QKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEA 215
Q EQL R + + +H +LL++ E+ + ++LREKE E+E RKN EL R +++VEA
Sbjct: 144 QAEQLXRALAEKXXRHYRALLSTAEEAVARRLREKEAEVEMATRKNAELEARAAKLSVEA 323
Query: 216 QNWHYRAKYNESVVNALRNNLQQAI-SHGVEQ----GKEGFGDSEVDDAAS-YIDPNNFL 269
Q W +A+ E+ +L+ LQQ I SHG E+ G + + +DA S YIDP +
Sbjct: 324 QVWQAKARAQEATAASLQAQLQQTIMSHGGEELAAVGVSSAVEGQAEDAESAYIDPERVV 503
Query: 270 SIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIK 329
A K CR C + S++++PCRHL +C +CD CPVC +K
Sbjct: 504 VATTARPK-------------CRGCAKRVASVVVLPCRHLCICTECDAHFRACPVCLTLK 644
Query: 330 TASVEVYLS 338
++VEV+LS
Sbjct: 645 NSTVEVFLS 671
>TC228566 weakly similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana genomic
DNA, chromosome 5, TAC clone:K17O22
(AT5g45100/K17O22_9), partial (56%)
Length = 1081
Score = 118 bits (295), Expect = 4e-27
Identities = 88/309 (28%), Positives = 138/309 (44%), Gaps = 29/309 (9%)
Frame = +3
Query: 59 PNKRSREMEDISKKQRLQISLN----YNVCQDDADWSASIPNPNALSTGLRLSYDDD--- 111
P++ E I Q+LQ LN YN D S+++P P + L Y +
Sbjct: 171 PSQLLTNRELIKSNQQLQHQLNSDYMYNTTTQ-MDSSSALPQPATMPESLLSFYQSNFCD 347
Query: 112 ----------------ERNSSVTSASGSMTAPPSIILS-----LGDNIRTELDRQQEELD 150
+R+ T+ S+ A +S L I + QQ E+D
Sbjct: 348 PNKADSGLTYHIPLQRKRSRDFTTELTSLPAHQKNKISSDPSFLNQEILYQFQNQQSEID 527
Query: 151 HYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQ 210
+ E++ + + K + +++I++ + KKL+EK++EI+ M + N L ER+K
Sbjct: 528 RVLAHHTEKVRMELEEQKMRQSRMFVSAIQEAMAKKLKEKDQEIQRMGKLNWTLQERVKS 707
Query: 211 VAVEAQNWHYRAKYNESVVNALRNNLQQAISH-GVEQGKEGFGDSEVDDAASYIDPNNFL 269
+ +E Q W A+ NES N LR+NL+Q ++H G E+ G DDA S N+
Sbjct: 708 LCMENQIWRELAQTNESTANYLRSNLEQVLAHVGEERATXG------DDAQSSCGSNDAA 869
Query: 270 SIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIK 329
S +G C+ C ++ +LL+PCRHL LC C + CP+C
Sbjct: 870 EAGNDTAASAAATGRGR---LCKNCGLRESVVLLLPCRHLCLCTMCGSTVRNCPICDSDM 1040
Query: 330 TASVEVYLS 338
ASV V LS
Sbjct: 1041DASVHVNLS 1067
>TC218572 similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K17O22 (AT5g45100/K17O22_9),
partial (43%)
Length = 716
Score = 114 bits (285), Expect = 6e-26
Identities = 65/200 (32%), Positives = 102/200 (50%), Gaps = 6/200 (3%)
Frame = +1
Query: 145 QQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
QQ E+D +I E++ + + + + L+ +I++ + KKL+EK+EEI+ + + N L
Sbjct: 34 QQSEIDRFIAQHTEKVRMELEEQRVRQSRMLIAAIQEAVAKKLKEKDEEIQRVGKLNWVL 213
Query: 205 AERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYID 264
ER+K + VE Q W A+ NE+ N LRNNL+Q ++H E + V+ A S
Sbjct: 214 QERVKNLCVENQIWKELAQTNEATANNLRNNLEQVLAHVSEDHHHNLHHTTVEAAESSCA 393
Query: 265 PNNFLSIPGAPMKSIHPRYQGMEN------LSCRACKAKDVSMLLIPCRHLSLCKDCDGF 318
NN S + G ++ C C ++ +LL+PCRHL LC C+
Sbjct: 394 SNNNNSHHREEEEVCGGSGNGKQSDGVLGKRMCNQCGVRESIVLLLPCRHLCLCTMCEST 573
Query: 319 INVCPVCQMIKTASVEVYLS 338
+ CP+CQ ASV V S
Sbjct: 574 VRNCPLCQSGINASVHVNYS 633
>BI969751
Length = 734
Score = 107 bits (266), Expect = 1e-23
Identities = 59/198 (29%), Positives = 102/198 (50%), Gaps = 5/198 (2%)
Frame = -1
Query: 145 QQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
QQ E+D +I+ E+L +++ + H+A LL +E + LR+K+EEI +K+ EL
Sbjct: 620 QQREIDXHIRSHNEKLRILLQEQRXXHVAELLKKVESNALHLLRQKDEEIAQATKKSTEL 441
Query: 205 AERIKQVAVEAQNWHYRAKYNESVVNALRNNL----QQAISHGVEQGKEGFGDSEVDDAA 260
E + ++ VE Q+W A+ NE++V +L N L ++A+ ++ E D + + A
Sbjct: 440 KEFMTRLEVENQSWRKVAEENEAMVLSLHNTLEDMKERALYRVTKEDAESCCDENMRNRA 261
Query: 261 SYIDPNNFLSIPGAPMKSIHPRYQ-GMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFI 319
G + Q + C+ C +++ + +PCRHL CK C+ F+
Sbjct: 260 MEEGTGENRLCGGGGAGGVEEVEQIRKRTMDCKCCNSQNSCFMFLPCRHLCSCKTCEPFL 81
Query: 320 NVCPVCQMIKTASVEVYL 337
VCPVC M K +S+E +
Sbjct: 80 QVCPVCSMPKKSSIETLI 27
>BM892823 similar to GP|22946353|gb CG6214-PA {Drosophila melanogaster},
partial (0%)
Length = 421
Score = 94.4 bits (233), Expect = 7e-20
Identities = 44/54 (81%), Positives = 48/54 (88%)
Frame = +3
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNATNQLQLFGNLQAGRNVDPVSYIGNEHIS 54
M GGNNGNRLLPVFLDE QF+ QTN +NQLQLFGNLQAG +VDPV+Y GNEHIS
Sbjct: 258 MLGGNNGNRLLPVFLDETQFRHQTNVSNQLQLFGNLQAGCSVDPVNYFGNEHIS 419
>TC230232 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-like protein,
partial (27%)
Length = 939
Score = 83.6 bits (205), Expect = 1e-16
Identities = 45/136 (33%), Positives = 78/136 (57%), Gaps = 8/136 (5%)
Frame = +1
Query: 134 LGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEE 193
LG ++ ++ +QQ +++H I + E++ + + +++ ++ +IE G++KKL+ KEEE
Sbjct: 418 LGQDVSLQIQQQQLDIEHLIMQRMEKVRMEIDEKRKRQARRIIEAIEVGVMKKLKTKEEE 597
Query: 194 IENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQ--------AISHGVE 245
IE + + N L E++K + +E Q W A+ NE+ NALR NL+Q A V
Sbjct: 598 IEKIGKLNWALEEKVKHLCMENQVWRNIAETNEATANALRCNLEQVLAQRGGMAAEEDVG 777
Query: 246 QGKEGFGDSEVDDAAS 261
G G +E+DDA S
Sbjct: 778 GGATVCGGAEMDDAES 825
>TC230231 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-like protein,
partial (17%)
Length = 615
Score = 80.5 bits (197), Expect = 1e-15
Identities = 49/169 (28%), Positives = 80/169 (46%), Gaps = 18/169 (10%)
Frame = +2
Query: 188 REKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISH----- 242
+ KEEEIE + + N + + + Q W A E+ NA+R NL+Q ++
Sbjct: 2 KTKEEEIEIIGKMNWAXXXXVMHLCMXNQVWRNIAXTKEATANAMRCNLEQVLAQRGGMA 181
Query: 243 ---GVEQGKEGFGDSEVDDAASYI----------DPNNFLSIPGAPMKSIHPRYQGMENL 289
V G G +E+DDA S + + ++ G + + G
Sbjct: 182 SEEDVGVGATVCGGAEMDDAESCCGSTEEDGLEKETGGWRTLAGCA--GVKDKEGGGNGR 355
Query: 290 SCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
CR C+ ++ +L++PCRHL LC C +++CP+C+ KTASV V +S
Sbjct: 356 LCRNCRKEESCVLILPCRHLCLCTVCGSSLHICPICKSYKTASVHVNMS 502
>AW704674
Length = 411
Score = 65.9 bits (159), Expect = 3e-11
Identities = 31/85 (36%), Positives = 55/85 (64%)
Frame = +3
Query: 145 QQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
QQ E+DH+I+ E+L +++ +++H+A LL +E + LR+K+EEI +K+ EL
Sbjct: 156 QQREIDHHIRSHNEKLRILLQEQRKQHVAELLKKVESNALHLLRQKDEEIAQATKKSTEL 335
Query: 205 AERIKQVAVEAQNWHYRAKYNESVV 229
E + ++ VE Q+W A+ NE++V
Sbjct: 336 KEFMTRLEVENQSWRKVAEENEAMV 410
>TC208035 similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (15%)
Length = 445
Score = 58.2 bits (139), Expect = 5e-09
Identities = 20/45 (44%), Positives = 32/45 (70%)
Frame = +2
Query: 294 CKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
C + S++++PCRHL +C +CDG CPVC +K ++++VYLS
Sbjct: 44 CAKRVASVVVLPCRHLCICAECDGHFRACPVCLTVKNSTIQVYLS 178
>AI759807
Length = 207
Score = 50.1 bits (118), Expect = 1e-06
Identities = 17/33 (51%), Positives = 23/33 (69%)
Frame = +1
Query: 305 PCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
PCRHL +C +CDG CPVC +K +++VYL
Sbjct: 109 PCRHLCICAECDGHFRACPVCLTVKNLTIQVYL 207
>BE346744 weakly similar to GP|21593126|gb| inhibitor of apoptosis-like
protein {Arabidopsis thaliana}, partial (13%)
Length = 145
Score = 47.0 bits (110), Expect = 1e-05
Identities = 20/45 (44%), Positives = 27/45 (59%)
Frame = -1
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEV 335
C C ++ +LL+PCRHL LC C ++ CP+CQ ASV V
Sbjct: 136 CNQCGVRESIVLLLPCRHLCLCTMCWSTVHNCPLCQCGINASVHV 2
>TC217510
Length = 791
Score = 44.3 bits (103), Expect = 8e-05
Identities = 17/50 (34%), Positives = 27/50 (54%)
Frame = +1
Query: 287 ENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
E + CR C +++++L+PCRH LC C CP+C+ + VY
Sbjct: 322 EKVLCRVCFEGEINVVLLPCRHRVLCSTCSEKCKKCPICRDSIAERLPVY 471
>TC230260
Length = 783
Score = 43.5 bits (101), Expect = 1e-04
Identities = 14/46 (30%), Positives = 29/46 (62%)
Frame = +3
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
CR C + +V + ++PC H+ LC+ C ++ CP C++ T ++ ++
Sbjct: 477 CRVCLSSEVDITIVPCGHV-LCRRCSSAVSRCPFCRLQVTKAIRIF 611
>TC231131 similar to GB|BAB71852.1|16902294|AB062739 kinesin-related protein
{Arabidopsis thaliana;} , partial (15%)
Length = 725
Score = 43.1 bits (100), Expect = 2e-04
Identities = 14/36 (38%), Positives = 21/36 (57%)
Frame = +3
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
C+ C + +L+PCRH LCK C + CP+C+
Sbjct: 486 CKVCFQSSTAAILLPCRHFCLCKSCSLACSECPICR 593
>TC208814
Length = 638
Score = 42.7 bits (99), Expect = 2e-04
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +1
Query: 287 ENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
E + CR C + ++++L+PCRH LC C CPVC+
Sbjct: 331 EKILCRICFEEQINVVLLPCRHHILCSTCCEKCKRCPVCR 450
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,852,863
Number of Sequences: 63676
Number of extensions: 180663
Number of successful extensions: 1123
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 1106
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1110
length of query: 338
length of database: 12,639,632
effective HSP length: 98
effective length of query: 240
effective length of database: 6,399,384
effective search space: 1535852160
effective search space used: 1535852160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0008.8


