

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
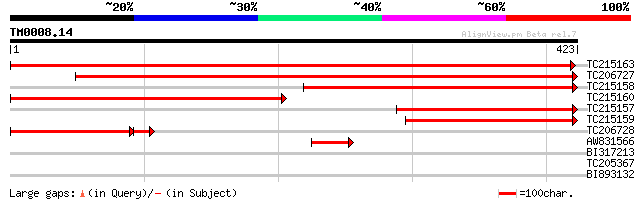
Query= TM0008.14
(423 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215163 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-sub... 789 0.0
TC206727 similar to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subun... 637 0.0
TC215158 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-sub... 384 e-107
TC215160 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-sub... 378 e-105
TC215157 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-sub... 261 3e-70
TC215159 homologue to UP|Q9SGY2 (Q9SGY2) F20B24.11 (ATP-citrate ... 240 8e-64
TC206728 similar to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subun... 142 2e-38
AW831566 similar to GP|15919089|emb ATP citrate lyase b-subunit ... 58 9e-09
BI317213 31 1.2
TC205367 similar to UP|O24525 (O24525) Glutathione-conjugate tra... 30 2.7
BI893132 GP|22832616|g CG11940-PB {Drosophila melanogaster}, par... 29 4.6
>TC215163 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
complete
Length = 1654
Score = 789 bits (2037), Expect = 0.0
Identities = 392/422 (92%), Positives = 410/422 (96%)
Frame = +1
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRL+KEHFKRLSGKELPIKSAQ+T STDF+ELQ+KEPWLSSSKLVVKPD
Sbjct: 271 MARKKIREYDSKRLLKEHFKRLSGKELPIKSAQITASTDFTELQEKEPWLSSSKLVVKPD 450
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVALNLDFAQV SFVKERLGKEVEM GCKGPITTFIVEPFIPHNEEFYLN
Sbjct: 451 MLFGKRGKSGLVALNLDFAQVVSFVKERLGKEVEMSGCKGPITTFIVEPFIPHNEEFYLN 630
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVSERLGNSISFSECGGI+IE+NWDKVKT F+PTG+SLTSEIVAPLVATLPLEIKGEIEE
Sbjct: 631 IVSERLGNSISFSECGGIEIEDNWDKVKTAFVPTGMSLTSEIVAPLVATLPLEIKGEIEE 810
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
FLKV+FTLFQDLDFTFLEMNPFTLVDGKP PLDMRGELDDTAAFKNFKKWG+IEFPLPFG
Sbjct: 811 FLKVVFTLFQDLDFTFLEMNPFTLVDGKPYPLDMRGELDDTAAFKNFKKWGDIEFPLPFG 990
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RVMSATESF+H LDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN
Sbjct: 991 RVMSATESFIHGLDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 1170
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAPKE+EVLQYARVVIDCATANPD QKRALVIGGGIANFTDVAATFSGIIRALKE
Sbjct: 1171YAEYSGAPKEEEVLQYARVVIDCATANPDDQKRALVIGGGIANFTDVAATFSGIIRALKE 1350
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCIT 420
KESKLKAARMHI+VRRGGPNYQKGLALMRALG++IGIP EVYGP+ATM GICKEA+Q +
Sbjct: 1351KESKLKAARMHIFVRRGGPNYQKGLALMRALGKDIGIPNEVYGPKATMFGICKEAMQFMN 1530
Query: 421 AA 422
A
Sbjct: 1531TA 1536
>TC206727 similar to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (85%)
Length = 1389
Score = 637 bits (1642), Expect = 0.0
Identities = 308/374 (82%), Positives = 345/374 (91%)
Frame = +2
Query: 50 LSSSKLVVKPDMLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEP 109
LSS++LVVKPDMLFGKRGKSGLVALNLD AQVA FVK RLG EVEMGGCK PITTFIVEP
Sbjct: 2 LSSTRLVVKPDMLFGKRGKSGLVALNLDIAQVAEFVKARLGVEVEMGGCKAPITTFIVEP 181
Query: 110 FIPHNEEFYLNIVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVAT 169
F+PH++EFYL+IVSERLG++ISFSECGGI+IEENWDKVKT+F+PT LT E APL+A
Sbjct: 182 FVPHDQEFYLSIVSERLGSTISFSECGGIEIEENWDKVKTIFLPTEKPLTPEACAPLIAI 361
Query: 170 LPLEIKGEIEEFLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKK 229
LPLEI+G I +F+ +F +F+DLDF+FLEMNPFTLV+ KP PLDMRGELDDTAAFKNF K
Sbjct: 362 LPLEIRGTIGDFIMGVFAVFKDLDFSFLEMNPFTLVNEKPYPLDMRGELDDTAAFKNFNK 541
Query: 230 WGNIEFPLPFGRVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTV 289
WGNIEFPLPFGR++S TESF+H LD+KTSASLKFT+LNPKGRIWTMVAGGGASVIYADTV
Sbjct: 542 WGNIEFPLPFGRILSPTESFIHSLDDKTSASLKFTILNPKGRIWTMVAGGGASVIYADTV 721
Query: 290 GDLGFASELGNYAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAA 349
GDLG+ASELGNYAEYSGAP E+EVLQYARVVIDCAT +PDG+KRAL+IGGGIANFTDVAA
Sbjct: 722 GDLGYASELGNYAEYSGAPNEEEVLQYARVVIDCATEDPDGRKRALLIGGGIANFTDVAA 901
Query: 350 TFSGIIRALKEKESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMT 409
TF+GIIRALKEKESKLKAA+MHIYVRRGGPNYQ GLA MRALGEE+G+PI+VYGPEATMT
Sbjct: 902 TFNGIIRALKEKESKLKAAQMHIYVRRGGPNYQTGLAKMRALGEELGVPIQVYGPEATMT 1081
Query: 410 GICKEAIQCITAAA 423
GICK+AI CI + A
Sbjct: 1082GICKQAIDCIMSEA 1123
>TC215158 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (48%)
Length = 1017
Score = 384 bits (986), Expect = e-107
Identities = 191/204 (93%), Positives = 196/204 (95%)
Frame = +3
Query: 220 DTAAFKNFKKWGNIEFPLPFGRVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGG 279
DTAAFKNFKKWGNIEFPLPFGRVMS TE+FVH LDEKTSASLKFTVLNP GRIWTMVAGG
Sbjct: 3 DTAAFKNFKKWGNIEFPLPFGRVMSTTEAFVHGLDEKTSASLKFTVLNPMGRIWTMVAGG 182
Query: 280 GASVIYADTVGDLGFASELGNYAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGG 339
GASVIYADTVGDLG+A ELGNYAEYSGAPKEDEVLQYARVVIDCAT+NPDGQKRALVIGG
Sbjct: 183 GASVIYADTVGDLGYAPELGNYAEYSGAPKEDEVLQYARVVIDCATSNPDGQKRALVIGG 362
Query: 340 GIANFTDVAATFSGIIRALKEKESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPI 399
GIANFTDVAATFSGIIRALKEKE KLK A+MHIYVRRGGPNYQKGLA MRALGEEIGIPI
Sbjct: 363 GIANFTDVAATFSGIIRALKEKEQKLKEAKMHIYVRRGGPNYQKGLAKMRALGEEIGIPI 542
Query: 400 EVYGPEATMTGICKEAIQCITAAA 423
EVYGPEATMTGICK+AIQ ITAAA
Sbjct: 543 EVYGPEATMTGICKQAIQYITAAA 614
>TC215160 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (48%)
Length = 739
Score = 378 bits (971), Expect = e-105
Identities = 189/206 (91%), Positives = 199/206 (95%)
Frame = +2
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRL+KEHFKR+SG+ELPIKSAQVT ST+FSEL +KEPWL SSKLVVKPD
Sbjct: 122 MARKKIREYDSKRLLKEHFKRISGQELPIKSAQVTESTNFSELAEKEPWLLSSKLVVKPD 301
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVALNLD A+V SFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN
Sbjct: 302 MLFGKRGKSGLVALNLDLAEVDSFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 481
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVSERLGNSISFSECGGI+IEENWDKVKTVF+PTGVSLTSE +APLVATLPLEIKGEIEE
Sbjct: 482 IVSERLGNSISFSECGGIEIEENWDKVKTVFMPTGVSLTSESIAPLVATLPLEIKGEIEE 661
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVD 206
FLKVIFTLFQDLD TFLEMNPFTLV+
Sbjct: 662 FLKVIFTLFQDLDXTFLEMNPFTLVN 739
>TC215157 homologue to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (32%)
Length = 689
Score = 261 bits (668), Expect = 3e-70
Identities = 132/135 (97%), Positives = 134/135 (98%)
Frame = +2
Query: 289 VGDLGFASELGNYAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVA 348
VGDLGFASELGNYAEYSGAPKE+EVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVA
Sbjct: 8 VGDLGFASELGNYAEYSGAPKEEEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVA 187
Query: 349 ATFSGIIRALKEKESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATM 408
ATFSGIIRALKEKESKLKAARMHI+VRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATM
Sbjct: 188 ATFSGIIRALKEKESKLKAARMHIFVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATM 367
Query: 409 TGICKEAIQCITAAA 423
TGICKEAIQ ITAAA
Sbjct: 368 TGICKEAIQFITAAA 412
>TC215159 homologue to UP|Q9SGY2 (Q9SGY2) F20B24.11 (ATP-citrate lyase
subunit A) (At1g10670/F20B24_11), partial (30%)
Length = 601
Score = 240 bits (613), Expect = 8e-64
Identities = 121/128 (94%), Positives = 125/128 (97%)
Frame = +2
Query: 296 SELGNYAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGII 355
SELGNYAEY GAP+E+EVLQYARVVIDCATANPD QKRALVIGGGIANFTDVAATFSGII
Sbjct: 2 SELGNYAEYRGAPQEEEVLQYARVVIDCATANPDDQKRALVIGGGIANFTDVAATFSGII 181
Query: 356 RALKEKESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEA 415
RALKEKESKLKAARMHI+VRRGGPNYQKGLALMRALGE+IGIPIEVYGPEATMTGICKEA
Sbjct: 182 RALKEKESKLKAARMHIFVRRGGPNYQKGLALMRALGEDIGIPIEVYGPEATMTGICKEA 361
Query: 416 IQCITAAA 423
IQ ITAAA
Sbjct: 362 IQFITAAA 385
>TC206728 similar to UP|Q93YH3 (Q93YH3) ATP citrate lyase b-subunit ,
partial (26%)
Length = 437
Score = 142 bits (357), Expect(2) = 2e-38
Identities = 73/93 (78%), Positives = 80/93 (85%)
Frame = +1
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREY SKRL+KEH KRL+ +L I SAQVT STDF+EL D+ PWLSS++LVVKPD
Sbjct: 34 MARKKIREYHSKRLLKEHLKRLASIDLQILSAQVTESTDFTELTDQHPWLSSTRLVVKPD 213
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEV 93
MLFGKRGKSGLVALNLD AQVA FVK RLG EV
Sbjct: 214 MLFGKRGKSGLVALNLDIAQVAEFVKARLGVEV 312
Score = 35.4 bits (80), Expect(2) = 2e-38
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = +3
Query: 93 VEMGGCKGPITTFIVE 108
VEMGGCK PITTFIVE
Sbjct: 390 VEMGGCKAPITTFIVE 437
>AW831566 similar to GP|15919089|emb ATP citrate lyase b-subunit {Lupinus
albus}, partial (6%)
Length = 388
Score = 57.8 bits (138), Expect = 9e-09
Identities = 25/31 (80%), Positives = 27/31 (86%)
Frame = +2
Query: 226 NFKKWGNIEFPLPFGRVMSATESFVHELDEK 256
N +WGNIEFPLPFGRVMS TE+FVH LDEK
Sbjct: 95 NICRWGNIEFPLPFGRVMSTTEAFVHGLDEK 187
>BI317213
Length = 343
Score = 30.8 bits (68), Expect = 1.2
Identities = 25/83 (30%), Positives = 36/83 (43%), Gaps = 2/83 (2%)
Frame = +2
Query: 9 YDSKRLVK--EHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPDMLFGKR 66
Y KR + EH +R++ + LP+ S+ + QD E WL SSKL+ + GK
Sbjct: 47 YCQKRFLAKWEHHQRVTYQSLPLMSSLRLMGLSLASQQDLE*WLLSSKLL*MQLEVCGKH 226
Query: 67 GKSGLVALNLDFAQVASFVKERL 89
L V V +RL
Sbjct: 227 NNLHPSRLEWSIXLVPRVVDKRL 295
>TC205367 similar to UP|O24525 (O24525) Glutathione-conjugate transporter
AtMRP4, partial (23%)
Length = 1548
Score = 29.6 bits (65), Expect = 2.7
Identities = 27/98 (27%), Positives = 41/98 (41%), Gaps = 17/98 (17%)
Frame = +3
Query: 196 FLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIE----------FP------LPF 239
FL+ +P +GK L + E+++ FK W N++ +P L
Sbjct: 390 FLKASP*ASTEGKRLELLVEQEVENQL*FKFSSGWWNLQEEK*SLTALTYPPWGFMILGH 569
Query: 240 GRVMSATE-SFVHELDEKTSASLKFTVLNPKGRIWTMV 276
G V S SF+ E E T L T + GR+W +V
Sbjct: 570 GLVSSLRSLSFLKEPSEVTLIQLDSTQMKRYGRVWNVV 683
>BI893132 GP|22832616|g CG11940-PB {Drosophila melanogaster}, partial (1%)
Length = 421
Score = 28.9 bits (63), Expect = 4.6
Identities = 16/72 (22%), Positives = 33/72 (45%), Gaps = 1/72 (1%)
Frame = +1
Query: 110 FIPHNEEFYLNIVSERLGNSISFSECG-GIDIEENWDKVKTVFIPTGVSLTSEIVAPLVA 168
F+PH + LNI RL + G G+ I + + + + S++ + + ++
Sbjct: 118 FVPHGHDDILNIAIGRLDHGGRVHAIGSGVTISQYYGRTSRASSSSSTSISQQQLVEIIG 297
Query: 169 TLPLEIKGEIEE 180
+L E + E +E
Sbjct: 298 SLKEEWRNEFKE 333
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,001,655
Number of Sequences: 63676
Number of extensions: 195332
Number of successful extensions: 789
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 787
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 787
length of query: 423
length of database: 12,639,632
effective HSP length: 100
effective length of query: 323
effective length of database: 6,272,032
effective search space: 2025866336
effective search space used: 2025866336
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0008.14


