

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
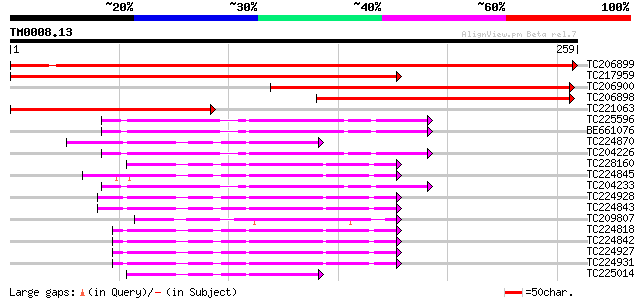
Query= TM0008.13
(259 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206899 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition partic... 398 e-111
TC217959 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition partic... 253 6e-68
TC206900 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition partic... 223 9e-59
TC206898 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition partic... 215 1e-56
TC221063 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition partic... 153 7e-38
TC225596 homologue to UP|O81695 (O81695) Ras-like small monomeri... 47 1e-05
BE661076 47 1e-05
TC224870 homologue to GB|AAB91395.1|2689631|AF022389 ADP-ribosyl... 46 2e-05
TC204226 homologue to UP|SARA_ARATH (O04834) GTP-binding protein... 45 4e-05
TC228160 homologue to GB|AAB17725.1|1654142|BCU38470 small GTP-b... 44 6e-05
TC224845 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor... 44 7e-05
TC204233 homologue to UP|SARA_ARATH (O04834) GTP-binding protein... 44 1e-04
TC224928 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor... 43 1e-04
TC224843 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor... 43 1e-04
TC209807 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor... 43 2e-04
TC224818 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete 42 2e-04
TC224842 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor... 42 2e-04
TC224927 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete 42 2e-04
TC224931 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor... 42 2e-04
TC225014 homologue to GB|AAB91395.1|2689631|AF022389 ADP-ribosyl... 41 5e-04
>TC206899 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (83%)
Length = 1006
Score = 398 bits (1023), Expect = e-111
Identities = 198/259 (76%), Positives = 222/259 (85%)
Frame = +1
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
M+ LE WK++ R W A + VPP QLY AA+ + TT+LLLS+R KRAK+NT+VL
Sbjct: 64 MEHLEQWKQQFLRWWRVA---VHTVPPIQLYAVAAVLILTTVLLLSIRFLKRAKSNTIVL 234
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSR 120
TGL+G+GKTVLFYQLRDGS H+GTVTSMEPNE TF+LHSET K KVKPV +VDVPGHSR
Sbjct: 235 TGLSGAGKTVLFYQLRDGSIHEGTVTSMEPNEDTFLLHSETVPKRKVKPVCVVDVPGHSR 414
Query: 121 LRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDK 180
LRPKLDEYLP+AA +VFVVDAVDFLPNCRAASEYLYDILTKGSVV+KKIPLLILCNKTDK
Sbjct: 415 LRPKLDEYLPKAAAIVFVVDAVDFLPNCRAASEYLYDILTKGSVVRKKIPLLILCNKTDK 594
Query: 181 VTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADAS 240
VTAH+KEFI +Q+ KEIDKLR SRSA+S AD+ NEF LGVPGE FSFTQC NKV DAS
Sbjct: 595 VTAHSKEFIGKQMGKEIDKLRESRSAISPADIANEFNLGVPGEPFSFTQCPNKVRLQDAS 774
Query: 241 GLTGEISQLEEFIREYVKP 259
GLTGEISQLE+FIREYVKP
Sbjct: 775 GLTGEISQLEQFIREYVKP 831
>TC217959 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (50%)
Length = 713
Score = 253 bits (646), Expect = 6e-68
Identities = 125/179 (69%), Positives = 148/179 (81%)
Frame = +3
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
M+ELE WKE+LS A D L +VPP QLY AAAI +FTTLLLLS+RLFKRAK+NTVVL
Sbjct: 177 MEELEQWKEQLSHFANLALDRLLEVPPNQLYAAAAIVIFTTLLLLSIRLFKRAKSNTVVL 356
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSR 120
GL+GSGKTV+FYQLRDGSTHQGTVTSMEPNEGTFILH+E T+KGK+KPVH+VDVPGHSR
Sbjct: 357 AGLSGSGKTVIFYQLRDGSTHQGTVTSMEPNEGTFILHNEKTRKGKIKPVHVVDVPGHSR 536
Query: 121 LRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
LRPKLDEYLP A +VFVVDA D +P CRAA+ YL + GS+V + +L++C ++D
Sbjct: 537 LRPKLDEYLPHTAVIVFVVDAKDSVPYCRAAA*YLIYLYIPGSIVSEYGAMLMVCWRSD 713
>TC206900 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (52%)
Length = 692
Score = 223 bits (567), Expect = 9e-59
Identities = 111/139 (79%), Positives = 122/139 (86%)
Frame = +1
Query: 120 RLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
RLRPKLDE AG+VFVVDA+DFLPNCRAASE LYD+LTKGSVV+KKIP+L CNKTD
Sbjct: 1 RLRPKLDEXXXXXAGIVFVVDALDFLPNCRAASEXLYDLLTKGSVVRKKIPVLXXCNKTD 180
Query: 180 KVTAHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADA 239
KV AHTKEFIR+Q+EKEI + RASRSA+S AD+ NEF LGVPGE F FTQC NKVTTADA
Sbjct: 181 KVPAHTKEFIRKQMEKEIGQFRASRSAISAADLANEFPLGVPGEPFFFTQCSNKVTTADA 360
Query: 240 SGLTGEISQLEEFIREYVK 258
SGLTGEISQLEEFIRE+VK
Sbjct: 361 SGLTGEISQLEEFIREHVK 417
>TC206898 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (44%)
Length = 669
Score = 215 bits (548), Expect = 1e-56
Identities = 105/118 (88%), Positives = 114/118 (95%)
Frame = +2
Query: 141 AVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKL 200
A+DFLPNCRAASEYLYD+LTKGSVV+KKIP+LILCNKTDKVTAHTKEFIR+Q+EKEIDKL
Sbjct: 2 ALDFLPNCRAASEYLYDLLTKGSVVRKKIPMLILCNKTDKVTAHTKEFIRKQMEKEIDKL 181
Query: 201 RASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFIREYVK 258
RASRSA+S AD+ NEFTLGVPGE FSFTQC NKVTTADASGLTGEISQLEEFIRE+VK
Sbjct: 182 RASRSAISAADIANEFTLGVPGEPFSFTQCSNKVTTADASGLTGEISQLEEFIREHVK 355
>TC221063 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (27%)
Length = 469
Score = 153 bits (387), Expect = 7e-38
Identities = 76/94 (80%), Positives = 84/94 (88%)
Frame = +2
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
M+ELE WKE+LS A D LR+VPP QLY AAAIA+FTTLLLLS+RLFKRAK+NT+VL
Sbjct: 188 MEELEQWKEQLSHFANLALDRLREVPPNQLYAAAAIAIFTTLLLLSIRLFKRAKSNTIVL 367
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGT 94
TGL+GSGKTVLFYQLRDGSTHQGTVTSMEPNEGT
Sbjct: 368 TGLSGSGKTVLFYQLRDGSTHQGTVTSMEPNEGT 469
>TC225596 homologue to UP|O81695 (O81695) Ras-like small monomeric
GTP-binding protein, complete
Length = 1089
Score = 46.6 bits (109), Expect = 1e-05
Identities = 46/151 (30%), Positives = 69/151 (45%)
Frame = +1
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 157 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQYPTSEELSI------ 312
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y Q VV++VDA D + E L +L+
Sbjct: 313 --GKIK-FKAFDLGGHQIARRVWKDYYAQVDAVVYLVDAYDKERFAESKKE-LDALLSDE 480
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P L+L NK D A ++E +R L
Sbjct: 481 SLA--SVPFLVLGNKIDIPYAASEEELRYHL 567
>BE661076
Length = 595
Score = 46.6 bits (109), Expect = 1e-05
Identities = 46/151 (30%), Positives = 69/151 (45%)
Frame = +2
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 95 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQYPTSEELSI------ 250
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y Q VV++VDA D + E L +L+
Sbjct: 251 --GKIK-FKAFDLGGHQIARRVWKDYYAQVDAVVYLVDAYDKERFAESKKE-LDALLSDE 418
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P L+L NK D A ++E +R L
Sbjct: 419 SLA--NVPFLVLGNKIDIPYAASEEELRYHL 505
>TC224870 homologue to GB|AAB91395.1|2689631|AF022389 ADP-ribosylation factor
{Vigna unguiculata;} , partial (76%)
Length = 601
Score = 45.8 bits (107), Expect = 2e-05
Identities = 32/117 (27%), Positives = 56/117 (47%)
Frame = +2
Query: 27 PEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVT 86
P Q+Y+ + + + L RLF + K +++ GL +GKT + Y+L+ G +
Sbjct: 155 PNQIYLFFKLKMGLSFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIV 316
Query: 87 SMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD 143
+ P G + ET + + + DV G ++RP Y G++FVVD+ D
Sbjct: 317 TTIPTIG---FNVETVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND 475
>TC204226 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 1062
Score = 44.7 bits (104), Expect = 4e-05
Identities = 45/151 (29%), Positives = 69/151 (44%)
Frame = +3
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 261 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 416
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D + E L +L+
Sbjct: 417 --GKIK-FKAFDLGGHQVARRVWKDYYAKVDAVVYLVDAFDKERFAESKKE-LDALLSDE 584
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P L+L NK D A ++E +R L
Sbjct: 585 SLA--NVPFLVLGNKIDIPYAASEEELRYHL 671
>TC228160 homologue to GB|AAB17725.1|1654142|BCU38470 small GTP-binding
protein ARF {Brassica rapa subsp. pekinensis;} ,
complete
Length = 1127
Score = 44.3 bits (103), Expect = 6e-05
Identities = 36/126 (28%), Positives = 59/126 (46%)
Frame = +2
Query: 54 KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV 113
K +++ GL +GKT + Y+L+ G V S P G + ET + +K +
Sbjct: 302 KEARILVLGLDNAGKTTILYRLQ-----MGEVVSTIPTIG---FNVETVQYNNIK-FQVW 454
Query: 114 DVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLI 173
D+ G + +RP Y P +++VVD+ D + A E + IL + + K +LI
Sbjct: 455 DLGGQTSIRPYWRCYFPNTQAIIYVVDSSD-VDRLVIAKEEFHAILEEEEL--KGAVVLI 625
Query: 174 LCNKTD 179
NK D
Sbjct: 626 FANKQD 643
>TC224845 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor 1-like
protein, complete
Length = 865
Score = 43.9 bits (102), Expect = 7e-05
Identities = 43/158 (27%), Positives = 71/158 (44%), Gaps = 12/158 (7%)
Frame = +2
Query: 34 AAIAVFTTLLLLSL---------RLFKRA---KTNTVVLTGLTGSGKTVLFYQLRDGSTH 81
A I + +LLLL L +LF R K +++ GL +GKT + Y+L+
Sbjct: 32 ANILILLSLLLLLLN*EMGLSFTKLFSRLFAKKEMRILMVGLDAAGKTTILYKLK----- 196
Query: 82 QGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDA 141
G + + P G + ET + + + DV G ++RP Y G++FVVD+
Sbjct: 197 LGEIVTTIPTIG---FNVETVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDS 364
Query: 142 VDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTD 179
D A + L+ +L + + + LL+ NK D
Sbjct: 365 ND-RDRVVEARDELHRMLNEDEL--RDAVLLVFANKQD 469
>TC204233 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 991
Score = 43.5 bits (101), Expect = 1e-04
Identities = 44/151 (29%), Positives = 69/151 (45%)
Frame = +3
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 168 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 323
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D + E L +L+
Sbjct: 324 --GKIK-FKAFDLGGHQVARRVWKDYYAKVDAVVYLVDAFDKERFAESKKE-LDALLSDE 491
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P L+L NK D A +++ +R L
Sbjct: 492 SLA--NVPFLVLGNKIDIPYAASEDELRYHL 578
>TC224928 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor, partial
(48%)
Length = 913
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +2
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 113 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 265
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 266 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 439
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 440 EDEL--RDAVLLVFANKQD 490
>TC224843 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor, partial
(48%)
Length = 1021
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +2
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 113 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 265
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 266 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 439
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 440 EDEL--RDAVLLVFANKQD 490
>TC209807 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor-like
protein, complete
Length = 796
Score = 42.7 bits (99), Expect = 2e-04
Identities = 39/127 (30%), Positives = 58/127 (44%), Gaps = 5/127 (3%)
Frame = +1
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPV--HIVDV 115
VV+ GL +GKT + Y+L H G V S P G + +K + K V + DV
Sbjct: 133 VVMLGLDAAGKTTILYKL-----HIGEVLSTVPTIGFNV------EKVQYKNVIFTVWDV 279
Query: 116 PGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEY---LYDILTKGSVVKKKIPLL 172
G +LRP Y G+++VVD++D +A E+ + D SV+ L
Sbjct: 280 GGQEKLRPLWRHYFNNTDGLIYVVDSLDRERIGKAKQEFQTIINDPFMLNSVI------L 441
Query: 173 ILCNKTD 179
+ NK D
Sbjct: 442 VFANKQD 462
>TC224818 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete
Length = 1067
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +1
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 157 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 309
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 310 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 477
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 478 DAVLLVFANKQD 513
>TC224842 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor 1-like
protein, complete
Length = 854
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +1
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 115 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 267
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 268 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 435
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 436 DAVLLVFANKQD 471
>TC224927 UP|Q6S4R7 (Q6S4R7) ADP-ribosylation factor, complete
Length = 825
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +1
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 112 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 264
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 265 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 432
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 433 DAVLLVFANKQD 468
>TC224931 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor 1-like
protein, complete
Length = 1014
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +3
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 252 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 404
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 405 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 572
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 573 DAVLLVFANKQD 608
>TC225014 homologue to GB|AAB91395.1|2689631|AF022389 ADP-ribosylation factor
{Vigna unguiculata;} , partial (60%)
Length = 444
Score = 41.2 bits (95), Expect = 5e-04
Identities = 26/90 (28%), Positives = 44/90 (48%)
Frame = +2
Query: 54 KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV 113
K T+++ GL +GKT + Y+L+ G + + P G + ET + + +
Sbjct: 41 KEMTILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNIS-FTVW 193
Query: 114 DVPGHSRLRPKLDEYLPQAAGVVFVVDAVD 143
DV G ++RP Y G++FVVD+ D
Sbjct: 194 DVGGQDKIRPLWRHYFQNTQGLIFVVDSND 283
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,821,673
Number of Sequences: 63676
Number of extensions: 133898
Number of successful extensions: 653
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 648
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 652
length of query: 259
length of database: 12,639,632
effective HSP length: 95
effective length of query: 164
effective length of database: 6,590,412
effective search space: 1080827568
effective search space used: 1080827568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0008.13


