

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
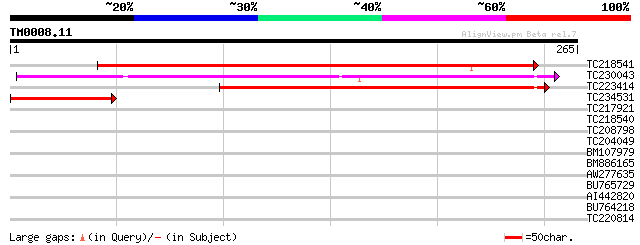
Query= TM0008.11
(265 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218541 weakly similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial... 298 2e-81
TC230043 weakly similar to GB|AAL31204.1|16974602|AY060578 AT3g2... 214 3e-56
TC223414 similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial (54%) 149 1e-36
TC234531 weakly similar to UP|Q9SGY3 (Q9SGY3) F20B24.10, partial... 90 9e-19
TC217921 similar to UP|Q8S8R0 (Q8S8R0) Predicted protein, partia... 36 0.016
TC218540 similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial (25%) 34 0.077
TC208798 similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_200, par... 29 1.9
TC204049 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to ch... 28 3.2
BM107979 28 3.2
BM886165 28 5.5
AW277635 similar to GP|20198043|gb| 26S proteasome regulatory su... 28 5.5
BU765729 28 5.5
AI442820 28 5.5
BU764218 similar to GP|7767671|gb|A F27F5.22 {Arabidopsis thalia... 27 9.5
TC220814 similar to UP|Q7Y231 (Q7Y231) At1g45150, partial (36%) 27 9.5
>TC218541 weakly similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial (46%)
Length = 1107
Score = 298 bits (762), Expect = 2e-81
Identities = 142/207 (68%), Positives = 168/207 (80%), Gaps = 1/207 (0%)
Frame = +3
Query: 42 KSRDERGERQQQRAGLLYEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVG 101
KSR+ER ERQ++ AG LYE E WNTCLK IHP WLLAYRI+SF+VL LL N+VADG G
Sbjct: 3 KSRNERRERQRETAGSLYEYEAWNTCLKGIHPAWLLAYRIISFLVLFSLLTANVVADGGG 182
Query: 102 ILYFYTQWTFTLVTIYFGLASFFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGD 161
I YFYTQWTFTLVTIYFGL S SIYGC +KH+K + TV A L+TE GTYVAPT+DG
Sbjct: 183 IFYFYTQWTFTLVTIYFGLGSCVSIYGCRYKHNKIDCTTVNRADLDTEEGTYVAPTLDGT 362
Query: 162 TDIPHMYKSTDAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYP-FLAPKD 220
++P++YK+++A QEP+T+NTAG WGYIFQI FQTCAGAVVLTD+VFWLVLYP +L KD
Sbjct: 363 PELPNLYKNSNANQEPYTRNTAGVWGYIFQITFQTCAGAVVLTDVVFWLVLYPTYLNTKD 542
Query: 221 FRLGLFAVCMHSVNAVFLLGETSLNGM 247
F L VC+HS+NA+FLLG+ SLN M
Sbjct: 543 FHLHFMDVCLHSLNAIFLLGDASLNCM 623
Score = 34.3 bits (77), Expect = 0.059
Identities = 13/17 (76%), Positives = 16/17 (93%)
Frame = +3
Query: 248 VKYLWLSRLFPGSCQVV 264
+K+LWLS+LFPGSCQ V
Sbjct: 825 LKHLWLSKLFPGSCQFV 875
>TC230043 weakly similar to GB|AAL31204.1|16974602|AY060578
AT3g27760/MGF10_16 {Arabidopsis thaliana;} , partial
(48%)
Length = 1408
Score = 214 bits (545), Expect = 3e-56
Identities = 111/255 (43%), Positives = 153/255 (59%), Gaps = 1/255 (0%)
Frame = +3
Query: 4 DTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYEDEL 63
DTT LSYWLNWRF +CA+ ++ ++ L +IWK + K R +GE Q+ G L DE
Sbjct: 102 DTTTLSYWLNWRFYLCAVCVLLSIVLSFLVIWKDKGSRKFRSGKGENQED--GTLSGDEA 275
Query: 64 WNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASF 123
W LK IHPV LLA+R+++F LL L+ + +G I ++YTQWTFTLVTIYFG AS
Sbjct: 276 WKPFLKEIHPVCLLAFRVIAFSSLLASLVAKIHINGRAIFFYYTQWTFTLVTIYFGFAST 455
Query: 124 FSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGDT-DIPHMYKSTDAYQEPHTQNT 182
S YGC+ + N V A ++ E+G Y+ P + DT ++ M D E
Sbjct: 456 LSAYGCYRHNKSSTINNVNVARIDVEQGPYM-PFLHQDTSNLSRMEHLADPLAEIQKNQV 632
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
A W YI QI+FQ AGAV+LTD ++WL+++PFL +D+ V MH++N VFLLG+
Sbjct: 633 APIWSYILQILFQMNAGAVMLTDCIYWLIIFPFLTLRDYDFNFMTVNMHTLNVVFLLGDA 812
Query: 243 SLNGMVKYLWLSRLF 257
+LN +K W F
Sbjct: 813 ALN-CLKIHWFGMSF 854
>TC223414 similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial (54%)
Length = 819
Score = 149 bits (377), Expect = 1e-36
Identities = 74/154 (48%), Positives = 99/154 (64%)
Frame = +3
Query: 99 GVGILYFYTQWTFTLVTIYFGLASFFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTI 158
G I Y+YTQWTFT +TIYFGL S SIYGC+ H+K G+ VG+ + E+G Y A +
Sbjct: 3 GGSIFYYYTQWTFTSITIYFGLGSLLSIYGCYQHHNKATGDKVGNVDGDAEQGMYDASAL 182
Query: 159 DGDTDIPHMYKSTDAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAP 218
++ KS +E + AG WGY FQI+FQ AGAV+LTD VFW ++ PFL
Sbjct: 183 PQSSNPFDPEKSLGDPEEVLVRQHAGIWGYTFQIIFQINAGAVMLTDCVFWFIIVPFLTI 362
Query: 219 KDFRLGLFAVCMHSVNAVFLLGETSLNGMVKYLW 252
KD+ L V MHS+NAVFL+G+T+LN +++ W
Sbjct: 363 KDYNLNFLIVIMHSINAVFLIGDTALN-CLRFPW 461
>TC234531 weakly similar to UP|Q9SGY3 (Q9SGY3) F20B24.10, partial (7%)
Length = 457
Score = 90.1 bits (222), Expect = 9e-19
Identities = 41/50 (82%), Positives = 44/50 (88%)
Frame = +1
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGER 50
M DTTALSYWLNWRFS CAL+I+ TMGL SFLIWKYEEFNKSR+ER ER
Sbjct: 307 MAPDTTALSYWLNWRFSPCALFILLTMGLASFLIWKYEEFNKSRNERRER 456
>TC217921 similar to UP|Q8S8R0 (Q8S8R0) Predicted protein, partial (13%)
Length = 555
Score = 36.2 bits (82), Expect = 0.016
Identities = 16/31 (51%), Positives = 22/31 (70%), Gaps = 2/31 (6%)
Frame = +3
Query: 188 YIFQIMFQTCAGAVVLTDLVFWL--VLYPFL 216
+I +TCAGA +LTD+VFW+ V+ PFL
Sbjct: 36 WIIWCTLKTCAGAAILTDIVFWVWGVIVPFL 128
>TC218540 similar to UP|Q6NPD1 (Q6NPD1) At5g62960, partial (25%)
Length = 526
Score = 33.9 bits (76), Expect = 0.077
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +1
Query: 248 VKYLWLSRLFPGSCQVV 264
+K+LWLS+LFPGSCQ +
Sbjct: 328 LKHLWLSKLFPGSCQFI 378
>TC208798 similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_200, partial (21%)
Length = 1094
Score = 29.3 bits (64), Expect = 1.9
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Frame = +3
Query: 81 IVSFIVLLGL---LIGNMVADGVGILYFYTQWTFTLVTIYFG 119
+++ +V LGL ++ VA G+L Y +WTFT+ I FG
Sbjct: 255 LLAAVVSLGLALLIVPRQVAPWTGLLLMY-EWTFTIFFILFG 377
>TC204049 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to chromaffin
granule ATPase II homolog, partial (24%)
Length = 1234
Score = 28.5 bits (62), Expect = 3.2
Identities = 14/47 (29%), Positives = 25/47 (52%), Gaps = 1/47 (2%)
Frame = -1
Query: 203 LTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLG-ETSLNGMV 248
+T + W L P+L ++ + MH +N + ++G E SLN +V
Sbjct: 712 ITPCLIWCKLVPYLQQLCSTSAIYLISMHFLNNLIIIGEEPSLNPLV 572
>BM107979
Length = 540
Score = 28.5 bits (62), Expect = 3.2
Identities = 13/39 (33%), Positives = 21/39 (53%)
Frame = +1
Query: 55 AGLLYEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIG 93
AG+LY+ E T +R H W YR+ I+ + ++G
Sbjct: 418 AGILYDMEATTTVGRRQHLYWSSCYRLXFIIMAVPCVVG 534
>BM886165
Length = 437
Score = 27.7 bits (60), Expect = 5.5
Identities = 18/70 (25%), Positives = 29/70 (40%), Gaps = 7/70 (10%)
Frame = +1
Query: 51 QQQRAGLLYEDELWNTCLKRIHPVWLLAY---RIVSFIVLLGLLIGNMVADGVGILYFY- 106
+ + G Y +W T + +H +WL+ +V I +I + ILY Y
Sbjct: 112 RSREVGQSYITSVWTTFVAMVHALWLMIKIRPEVVPCISFFASIINFTMLQFEVILYCYC 291
Query: 107 ---TQWTFTL 113
QWT+ L
Sbjct: 292 PDTLQWTWNL 321
>AW277635 similar to GP|20198043|gb| 26S proteasome regulatory subunit S2
(RPN1) {Arabidopsis thaliana}, partial (6%)
Length = 440
Score = 27.7 bits (60), Expect = 5.5
Identities = 17/70 (24%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Frame = -1
Query: 36 KYEEFNKSRDERGERQQQRAGLL---YEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLI 92
+ FN +R R + + LL E L + +K + P+ + ++ +L+ LL+
Sbjct: 431 RQRNFNGLGTDRSHRARGSSNLLPYALECNLLQSLVKILKPLHIQLQLLLQNQILIELLL 252
Query: 93 GNMVADGVGI 102
GN + G+G+
Sbjct: 251 GNGIGVGIGV 222
>BU765729
Length = 425
Score = 27.7 bits (60), Expect = 5.5
Identities = 16/42 (38%), Positives = 22/42 (52%)
Frame = +1
Query: 205 DLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGETSLNG 246
+L W L FL P + LG+FA C+H + L G L+G
Sbjct: 178 ELFHWCFL--FLLPSNIFLGVFATCLHVE*KIRLKGV*FLSG 297
>AI442820
Length = 408
Score = 27.7 bits (60), Expect = 5.5
Identities = 9/27 (33%), Positives = 15/27 (55%)
Frame = -1
Query: 19 CALWIIFTMGLGSFLIWKYEEFNKSRD 45
C + + L SFL+W Y + N+ R+
Sbjct: 90 CGTICAYKLALNSFLVWHYYDLNRERE 10
>BU764218 similar to GP|7767671|gb|A F27F5.22 {Arabidopsis thaliana}, partial
(3%)
Length = 445
Score = 26.9 bits (58), Expect = 9.5
Identities = 9/23 (39%), Positives = 17/23 (73%)
Frame = -2
Query: 3 TDTTALSYWLNWRFSICALWIIF 25
+D+ +LS+ RF +C++W+IF
Sbjct: 435 SDSISLSHSTFTRFKLCSIWVIF 367
>TC220814 similar to UP|Q7Y231 (Q7Y231) At1g45150, partial (36%)
Length = 1213
Score = 26.9 bits (58), Expect = 9.5
Identities = 9/23 (39%), Positives = 17/23 (73%)
Frame = -2
Query: 3 TDTTALSYWLNWRFSICALWIIF 25
+D+ +LS+ RF +C++W+IF
Sbjct: 903 SDSISLSHSTFTRFKLCSIWVIF 835
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.143 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,293,512
Number of Sequences: 63676
Number of extensions: 230490
Number of successful extensions: 1684
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1673
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1682
length of query: 265
length of database: 12,639,632
effective HSP length: 96
effective length of query: 169
effective length of database: 6,526,736
effective search space: 1103018384
effective search space used: 1103018384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0008.11


