

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004b.4
(929 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
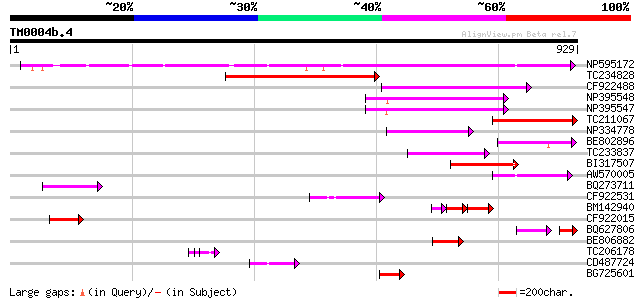
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 397 e-110
TC234828 234 2e-61
CF922488 160 2e-39
NP395548 reverse transcriptase [Glycine max] 159 7e-39
NP395547 reverse transcriptase [Glycine max] 147 2e-35
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 125 7e-29
NP334778 reverse transcriptase [Glycine max] 107 3e-23
BE802896 103 3e-22
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 98 1e-20
BI317507 97 3e-20
AW570005 94 3e-19
BQ273711 93 5e-19
CF922531 68 2e-11
BM142940 45 1e-10
CF922015 60 3e-09
BQ627806 45 1e-07
BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, pa... 50 6e-06
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 45 1e-04
CD487724 44 3e-04
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 44 3e-04
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 397 bits (1019), Expect = e-110
Identities = 289/953 (30%), Positives = 460/953 (47%), Gaps = 43/953 (4%)
Frame = +1
Query: 18 QVTNTAAREEAEAQRAAT----AALVAAQNQAEENARRV---------QREERELAAAQT 64
+ TN A E+ E ++ + L A +Q + + + ++E++ ++ Q
Sbjct: 106 EATNHAQMEKIEVMQSTNDSQFSQLNAVMSQVLQRLQNIPMSSHGASNSQKEQQRSSFQV 285
Query: 65 RGLN-DFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEAEYW 123
R + DF R FD + W+ + E+ F + T ++ + L + W
Sbjct: 286 RSVKLDFPR---------FDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPW 438
Query: 124 WRGARAMMEADHQAITWECFRGAFLDKYFPRSARAAKEAQFLRLRQGGMTVAEYAAKLES 183
++ M++ +W+ F A L+ F SA A +L Q TV EY + +
Sbjct: 439 YQ----MLQKTEPFSSWQAFTRA-LELDFGPSAYDCPRATLFKLNQSA-TVNEYYMQFTA 600
Query: 184 LAKHFRYFRGQIDEGYMCERFIEGLCYELQR---AVQPLGLNRYQVLVEKTKGIEAIDNA 240
L + + + F+ GL E+ R A++P L + L + +
Sbjct: 601 LVNRVDGLSAEA----ILDCFVSGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPK 768
Query: 241 RGKFQGLNKPYQGSGGPARTNQGRGDKGRHFQKKPYVRPQGRGTTSGSFYPTGGNA--IA 298
F L + + + + T + ++ KP + P ++ F N I+
Sbjct: 769 TKTFSNLARNF--TSNTSATQKYPPTNQKNDNPKPNLPPLLPTPSTKPFNLRNQNIKKIS 942
Query: 299 LRTPSGNREDVTCFRCNKKGHYANHCSESLAACWNCNKPGHTAAECRIPKVEAAANVAGA 358
RE C+ C++K A+ C + + ++ E A
Sbjct: 943 PAEIQLRREKNLCYFCDEKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMVTEEANMDDDT 1122
Query: 359 RRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDIACAARLKLEV 418
S++ G IR T ++ G ++ L D G++ +FI A LKL V
Sbjct: 1123 HH-------LSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPV 1281
Query: 419 SKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVIIGMDWLSH-- 476
P L V + L + P ++ + L + G DVI+G WL+
Sbjct: 1282 EPAP-NLRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLG 1458
Query: 477 HHVLLDCA--------NKVVIFPDAGLAEFLNS---YFSKLSLRKGA---------LSSL 516
HV A +K + G +E + +F +L K +
Sbjct: 1459 PHVADYAALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEV 1638
Query: 517 MSTTVVEAKENGVHGIAVV-QDFEDVFPEDVPG-IPPVRDMEFTIDIVPGTGPISIAPYR 574
T+ + N +A++ + VF VP +PP R+ + I + G+GP+ + PYR
Sbjct: 1639 PEDTLKDLPTNIDPELAILLHTYAQVFA--VPASLPPQREQDHAIPLKQGSGPVKVRPYR 1812
Query: 575 MAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLVKKKDGRSRLCVDYRQLNKVTIKNR 634
+ +++ ++++ +G I+PS SP+ P+LLVKKKDG R C DYR LN +T+K+
Sbjct: 1813 YPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDS 1992
Query: 635 YPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVT 694
+P+P +D+L+D+L GA FSK+DLRSGYHQI V+ ED +KTAFRT +GHYE+LVMPFG+T
Sbjct: 1993 FPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLT 2172
Query: 695 NAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANAT 754
NAPA F MN+IF L +FV+VF DDILIYS + ++H +HL VLQ L+ L+A +
Sbjct: 2173 NAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLS 2352
Query: 755 KCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGF 814
KC F EV +LGH +S G++++ +KV+AVL W P V +R F+GL GYYRRFI+ +
Sbjct: 2353 KCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSY 2532
Query: 815 AKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGL 874
A IAGPLT L +K+ F W + E +F +KK +T APVL+LP +P+ + DAS G+
Sbjct: 2533 ANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGV 2709
Query: 875 GCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVFALKIWRHYLYGSTFTV 927
G VL Q+ +AY S++L + + EL A+ AL +RHYL G+ F +
Sbjct: 2710 GAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFII 2868
>TC234828
Length = 857
Score = 234 bits (596), Expect = 2e-61
Identities = 121/254 (47%), Positives = 168/254 (65%), Gaps = 1/254 (0%)
Frame = +3
Query: 354 NVAGARRPTAGGRVYSISGTEAEEDDGLIRSTCEIAGNSLIALFDSGATHSFIDIACAAR 413
N++G R P RV+++SG+EA D LIR C IA L L+DSGATHSFI AC R
Sbjct: 99 NISGGR-PKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATHSFISHACVER 275
Query: 414 LKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKKFVANLICLPLKGLDVIIGMDW 473
L L ++LP+++ VSTP S+ + T+ CL+CP + + F+A+LICLPL LDVI+GMDW
Sbjct: 276 LGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLAHLDVILGMDW 455
Query: 474 LSHHHVLLDCANKVVIF-PDAGLAEFLNSYFSKLSLRKGALSSLMSTTVVEAKENGVHGI 532
LS +H+ LDC K+++F D +E L + ++ + VE ++ V I
Sbjct: 456 LSTNHIFLDCKEKMLVFGGDVVPSEPLKEDAANEETEDVRTYMVLFSMYVE-EDAEVSCI 632
Query: 533 AVVQDFEDVFPEDVPGIPPVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTK 592
VV +F +VFP+DV +PP R++EF ID+VPG P+SIAPYRM+P EL E+K+Q++DL
Sbjct: 633 PVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEVKAQVQDLLS 812
Query: 593 KGFIRPSVSPWGAP 606
K F+RPS SPWGAP
Sbjct: 813 KQFVRPSASPWGAP 854
>CF922488
Length = 741
Score = 160 bits (406), Expect = 2e-39
Identities = 91/245 (37%), Positives = 138/245 (56%)
Frame = +3
Query: 610 VKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKD 669
V K+DG+ +CVDYR LN + K+++PLP I+ L+D + FS +D SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 670 EDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRN 729
ED++KT F T +G + Y M FG+ N A + M +F + + + V++DD+++ SR
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 730 REEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWE 789
EEH +LR++ + LR L N KC F ++ K L + S* GI VD +KV+ +L
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 790 RPKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLT 849
+P T ++ F+G Y RFI PL L KNQ W DC +F+ +K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 850 TAPVL 854
VL
Sbjct: 723 NPHVL 737
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 159 bits (401), Expect = 7e-39
Identities = 92/254 (36%), Positives = 140/254 (54%), Gaps = 19/254 (7%)
Frame = +1
Query: 583 LKSQLEDLTKKGFIRP-SVSPWGAPVLLVKKKDGRS------------------RLCVDY 623
++ ++ L + G I P S S W +PVL+V KK+G + +LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 624 RQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGH 683
R+LN+ T K+ +PLP +D ++++L G A + +D GY+QI V +D +K AF +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 684 YEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQV 743
+ Y +PFG+ NAP F M IF +++ + VF+DD ++ + E + L VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 744 LRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGL 803
+ L N KC F + E LGH IS GI VD +K++ + P V IRSF+G
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 804 AGYYRRFIEGFAKI 817
A +YRRFI+ F K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 147 bits (371), Expect = 2e-35
Identities = 89/254 (35%), Positives = 132/254 (51%), Gaps = 19/254 (7%)
Frame = +1
Query: 583 LKSQLEDLTKKGFIRP-SVSPWGAPVLLVKKKDG------------------RSRLCVDY 623
++ ++ L + G I P S S W +PV +V KK G R R+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 624 RQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAFRTRYGH 683
R+LN+ T K+ YPLP +D ++ +L + + +D SGY+QI V +D +KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 684 YEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQV 743
+ Y MPFG+ NA F M IF +++ + VF+DD + + +L +VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 744 LRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPKTVTDIRSFIGL 803
L N KC F ++E LGH IS GI V K++ + P V I SF+G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 804 AGYYRRFIEGFAKI 817
G+YRRFI+ F K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 125 bits (315), Expect = 7e-29
Identities = 63/138 (45%), Positives = 87/138 (62%)
Frame = +1
Query: 792 KTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTA 851
K+V DIRSF GLA +YRRF+ F+ +A PL +L +KN F W E EQ+F +K++LT A
Sbjct: 106 KSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKA 285
Query: 852 PVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVVF 911
PVL LP + +E+ CDAS G+ VL+Q +AY S +L NYPT+D EL A++
Sbjct: 286 PVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYALIR 465
Query: 912 ALKIWRHYLYGSTFTVFS 929
A + W H+L F + S
Sbjct: 466 APQTWEHFLVCKEFVIHS 519
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 107 bits (266), Expect = 3e-23
Identities = 56/143 (39%), Positives = 85/143 (59%)
Frame = +3
Query: 618 RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGAAIFSKIDLRSGYHQIRVKDEDIQKTAF 677
R+CVDYR LN+ + K+ +PLP ID LM + A+FS +D SGY+QI++ ED++KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 678 RTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHL 737
T +G + Y VM FG+ N A + M +F + + + ++D+++ SR EEH +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 738 RQVLQVLRDKVLYANATKCEFWL 760
+ + LR L N KC F L
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>BE802896
Length = 416
Score = 103 bits (258), Expect = 3e-22
Identities = 55/134 (41%), Positives = 79/134 (58%), Gaps = 4/134 (2%)
Frame = -2
Query: 799 SFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTTAPVLTLPQ 858
SF+G AG+YRRFI F K+A PL+ L +K F + + C+ +F +K+ L T P++ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 859 EKEPYEVYCDASYQGLGCVLMQH----RKAVAYSSRQLKIHERNYPTHDLELAAVVFALK 914
P+E+ CDAS LG VL Q + + YSSR L + NY T + EL A+VFAL+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 915 IWRHYLYGSTFTVF 928
+ YL G+ V+
Sbjct: 55 KFHSYLLGTRIIVY 14
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 98.2 bits (243), Expect = 1e-20
Identities = 51/133 (38%), Positives = 80/133 (59%)
Frame = +2
Query: 653 FSKIDLRSGYHQIRVKDEDIQKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHPFL 712
FS +D SGY+QI + ED++KT F T +G + Y VM FG+ N A + M +FH +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 713 DRFVVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS* 772
+ + V++DD++ SR EH +L ++ L+ L N TKC F ++ K LG ++S
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 773 EGIAVDPSKVEAV 785
+GI +DP KV+A+
Sbjct: 362 KGIEIDPEKVKAL 400
>BI317507
Length = 359
Score = 97.4 bits (241), Expect = 3e-20
Identities = 53/113 (46%), Positives = 74/113 (64%), Gaps = 1/113 (0%)
Frame = -1
Query: 722 DILIYSRNREEHEEHLRQVLQVLRDKVLYANATKCEFWLEEVKFLGHVIS*EGIAVDPSK 781
+ILIYS + + H HL VL VL+ + L AN KC F +++LGHVIS + +A+D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 782 VEAVLAWERPKTVTDIRSFIGLAGYYRRFIEGFAKIA-GPLTKLTRKNQPFAW 833
V++V+ W PK V + SF+ L GYYR+FI+ + K+A PLT LT KN F W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLT-KNDGFKW 18
>AW570005
Length = 413
Score = 94.0 bits (232), Expect = 3e-19
Identities = 50/132 (37%), Positives = 75/132 (55%)
Frame = -2
Query: 791 PKTVTDIRSFIGLAGYYRRFIEGFAKIAGPLTKLTRKNQPFAWTEDCEQSFQDMKKRLTT 850
P+T +R F+ L G+YRRFI+G+A +A PL+ L K+ F W+ + + +FQ +K +T
Sbjct: 406 PRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVVTN 230
Query: 851 APVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAYSSRQLKIHERNYPTHDLELAAVV 910
VL LP +P+ V DAS +G VL Q +A+ S++ T+ ELAA+
Sbjct: 229 TLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAIT 50
Query: 911 FALKIWRHYLYG 922
+K WR YL G
Sbjct: 49 NVVKKWRQYLLG 14
>BQ273711
Length = 409
Score = 93.2 bits (230), Expect = 5e-19
Identities = 44/97 (45%), Positives = 58/97 (59%)
Frame = -2
Query: 55 EERELAAAQTRGLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETY 114
+ER++ A+ RGL F++ PPKFSG +DPE A LWL E EKIF + E KV Y T+
Sbjct: 291 QERDVEPAEYRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYATF 112
Query: 115 LLTGEAEYWWRGARAMMEADHQAITWECFRGAFLDKY 151
+L GEAE WW+ + A I W F+ FL+ Y
Sbjct: 111 MLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>CF922531
Length = 602
Score = 68.2 bits (165), Expect = 2e-11
Identities = 44/124 (35%), Positives = 72/124 (57%), Gaps = 1/124 (0%)
Frame = -2
Query: 492 DAGLAEFLNSYFSKLSLRKGALSSLMSTTVVEAKENGVHGIAVVQDFEDVFPEDVP-GIP 550
D +A L F L R+ +LS++ T+ E V + + +F D+FP+++P G+P
Sbjct: 364 DIKIALLLKQSFYLLLSRETSLSTVTIPTI-ETLPPKVQEL--LHEFGDIFPKEIPLGLP 194
Query: 551 PVRDMEFTIDIVPGTGPISIAPYRMAPAELTELKSQLEDLTKKGFIRPSVSPWGAPVLLV 610
P+R +E ID+VP + YR P E E++SQ+++L +KG+++ S+S VLLV
Sbjct: 193 PLRGIEHQIDLVPRASLPNRPTYRTNPQETKEIESQVKELLEKGWVQESLSLCVVLVLLV 14
Query: 611 KKKD 614
KKD
Sbjct: 13 PKKD 2
>BM142940
Length = 422
Score = 44.7 bits (104), Expect(3) = 1e-10
Identities = 18/42 (42%), Positives = 29/42 (68%)
Frame = -3
Query: 751 ANATKCEFWLEEVKFLGHVIS*EGIAVDPSKVEAVLAWERPK 792
AN KC F ++ ++L H+IS +G++ DP K+E ++AW PK
Sbjct: 156 ANRKKCSFGCKKFEYLVHMISVKGVSADPKKLEVMVAWPIPK 31
Score = 33.9 bits (76), Expect(3) = 1e-10
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = -1
Query: 716 VVVFIDDILIYSRNREEHEEHLRQVLQVLRDKVL 749
+ F DILIYS N +HE+HL+ VLQ ++ L
Sbjct: 260 ICFFSYDILIYSINEGKHEKHLKIVLQTFKEAKL 159
Score = 25.8 bits (55), Expect(3) = 1e-10
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = -2
Query: 691 FGVTNAPAVFMDYMNRIFHPFLDRF 715
F +TNA + F MN + PFL F
Sbjct: 421 FSLTNALSTFQALMNELLKPFLCNF 347
>CF922015
Length = 172
Score = 60.5 bits (145), Expect = 3e-09
Identities = 26/55 (47%), Positives = 37/55 (67%)
Frame = -3
Query: 66 GLNDFKRQDPPKFSGGFDPEEADLWLKELEKIFTFLRTTAEMKVDYETYLLTGEA 120
GL+ F+R +PP F GG+DPE A+ WL+E+EKIF + KV + T++L EA
Sbjct: 167 GLDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
>BQ627806
Length = 435
Score = 44.7 bits (104), Expect(2) = 1e-07
Identities = 21/57 (36%), Positives = 32/57 (55%)
Frame = +3
Query: 831 FAWTEDCEQSFQDMKKRLTTAPVLTLPQEKEPYEVYCDASYQGLGCVLMQHRKAVAY 887
F W E+ +++F +K L APVL LP + V DAS G+G +L Q+ +A+
Sbjct: 21 FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
Score = 30.4 bits (67), Expect(2) = 1e-07
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +1
Query: 901 THDLELAAVVFALKIWRHYLYGSTFTVFS 929
T+ ELAA+ A+K WR YL G F + +
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILT 318
>BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, partial (3%)
Length = 153
Score = 49.7 bits (117), Expect = 6e-06
Identities = 25/51 (49%), Positives = 31/51 (60%)
Frame = -1
Query: 693 VTNAPAVFMDYMNRIFHPFLDRFVVVFIDDILIYSRNREEHEEHLRQVLQV 743
+TNAP F MN IF L ++V+VF DDIL+YS EH HL V +V
Sbjct: 153 LTNAPTSFQCLMNHIFQHALRKYVLVFFDDILVYSSTWHEHLCHLEVVFKV 1
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 45.1 bits (105), Expect = 1e-04
Identities = 19/41 (46%), Positives = 23/41 (55%)
Frame = +2
Query: 304 GNREDVTCFRCNKKGHYANHCSESLAACWNCNKPGHTAAEC 344
G D C C + GHYA C ++A C NC PGH A+EC
Sbjct: 323 GFSRDNLCKNCKRPGHYARECP-NVAICHNCGLPGHIASEC 442
Score = 42.7 bits (99), Expect = 7e-04
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = +2
Query: 311 CFRCNKKGHYANHCSESLAACWNCNKPGHTAAEC 344
C C GH A+ C+ + CWNC +PGH A+ C
Sbjct: 401 CHNCGLPGHIASECTTK-SLCWNCKEPGHMASSC 499
Score = 42.7 bits (99), Expect = 7e-04
Identities = 20/52 (38%), Positives = 22/52 (41%)
Frame = +2
Query: 293 GGNAIALRTPSGNREDVTCFRCNKKGHYANHCSESLAACWNCNKPGHTAAEC 344
GG A G DV C C + GH + C L C NC GH A EC
Sbjct: 788 GGGGGARGGGGGGYRDVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 40.0 bits (92), Expect = 0.005
Identities = 19/47 (40%), Positives = 24/47 (50%)
Frame = +2
Query: 311 CFRCNKKGHYANHCSESLAACWNCNKPGHTAAECRIPKVEAAANVAG 357
C C K+GH A C+ AC NC K GH A +C + NV+G
Sbjct: 593 CNNCYKQGHIAAECTNE-KACNNCRKTGHLARDCPNDPICNLCNVSG 730
Score = 39.3 bits (90), Expect = 0.008
Identities = 19/40 (47%), Positives = 19/40 (47%), Gaps = 6/40 (15%)
Frame = +2
Query: 311 CFRCNKKGHYANHCSE------SLAACWNCNKPGHTAAEC 344
C C K GH A CS L C NC K GH AAEC
Sbjct: 515 CHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC 634
Score = 37.7 bits (86), Expect = 0.024
Identities = 15/37 (40%), Positives = 18/37 (48%)
Frame = +2
Query: 311 CFRCNKKGHYANHCSESLAACWNCNKPGHTAAECRIP 347
C+ C + GH A+ C C C K GH A EC P
Sbjct: 458 CWNCKEPGHMASSCPNE-GICHTCGKAGHRARECSAP 565
Score = 37.0 bits (84), Expect = 0.040
Identities = 23/55 (41%), Positives = 24/55 (42%)
Frame = +2
Query: 311 CFRCNKKGHYANHCSESLAACWNCNKPGHTAAECRIPKVEAAANVAGARRPTAGG 365
C C K GH A C C CN GH A +C PK ANV G R GG
Sbjct: 650 CNNCRKTGHLARDCPND-PICNLCNVSGHVARQC--PK----ANVLGDRSGGGGG 793
>CD487724
Length = 676
Score = 43.9 bits (102), Expect = 3e-04
Identities = 24/82 (29%), Positives = 40/82 (48%)
Frame = +3
Query: 393 LIALFDSGATHSFIDIACAARLKLEVSKLPFELTVSTPASKSLVTNTACLECPWMYLDKK 452
L+ L D G+TH+F+ ++L L P L V L T C P + +
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTP-PLRVMVGNGHHLKCTTICEAIPISIQNIE 200
Query: 453 FVANLICLPLKGLDVIIGMDWL 474
F+ +L LP+ G ++++G+ WL
Sbjct: 201 FLVHLYVLPIVGANIVLGVQWL 266
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 43.9 bits (102), Expect = 3e-04
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -3
Query: 607 VLLVKKKDGRSRLCVDYRQLNKVTIKNRYPLPRIDDLMDQL 647
V++VKK +G+ R+C DY LN K+ YPLP ID + D L
Sbjct: 124 VVMVKKPNGKWRICTDYIDLN*ACPKDAYPLPNIDHMTDGL 2
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,773,396
Number of Sequences: 63676
Number of extensions: 532048
Number of successful extensions: 2848
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 2740
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2817
length of query: 929
length of database: 12,639,632
effective HSP length: 106
effective length of query: 823
effective length of database: 5,889,976
effective search space: 4847450248
effective search space used: 4847450248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0004b.4


