

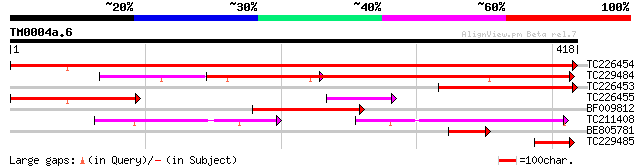
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.6
(418 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 764 0.0
TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 387 e-108
TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine c... 178 4e-45
TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine cytidy... 177 6e-45
BF009812 124 8e-29
TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine cyt... 114 6e-26
BE805781 59 5e-09
TC229485 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 46 3e-05
>TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (71%)
Length = 1889
Score = 764 bits (1972), Expect = 0.0
Identities = 373/420 (88%), Positives = 398/420 (93%), Gaps = 2/420 (0%)
Frame = +1
Query: 1 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFGGIG--HLPLPFPWSNSGIFHKK 58
MDYE+NSWIWEGV YYP +FGGLMVTAALLGLSTSYFGGIG +L LP WSN GIFHKK
Sbjct: 223 MDYENNSWIWEGVNYYPRVFGGLMVTAALLGLSTSYFGGIGVPYLSLPCSWSNLGIFHKK 402
Query: 59 KSGKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVVGLVNDEEILATKGPPVLTMAER 118
KSGKRR+RVYMDGCFDLMHYGHANALRQAKALGDELVVGLV+DEEI+A KGPPVL+M ER
Sbjct: 403 KSGKRRIRVYMDGCFDLMHYGHANALRQAKALGDELVVGLVSDEEIVANKGPPVLSMEER 582
Query: 119 LALVSGLKWVDEVITDAPYAITEQFLHRLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKA 178
LALVSGLKWVDEVITDAPYAITEQFL+RLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKA
Sbjct: 583 LALVSGLKWVDEVITDAPYAITEQFLNRLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKA 762
Query: 179 GRYKQIKRTEGVSSTDIVGRIMSSLKDQKTCEDQNGTDVKRQEESMSKDSHISQFLPTSR 238
GRYKQIKRTEGVSSTDIVGRI+SSL+DQK CED NGT+VK QEE+ S+ SHI+QFLPTSR
Sbjct: 763 GRYKQIKRTEGVSSTDIVGRILSSLRDQKNCEDHNGTEVKPQEENQSQASHIAQFLPTSR 942
Query: 239 RIVQFSNGKGPGPNARIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRG 298
RIVQFSNGKGPGPN+RIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRG
Sbjct: 943 RIVQFSNGKGPGPNSRIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRG 1122
Query: 299 NHYPIMHLHERSLSVLACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKSLHSERD 358
NHYPIMHLHERSLSVLACR+VDEVIIG+PWEIT DMITTFNIS+VVHGTVAEK+L+ E D
Sbjct: 1123NHYPIMHLHERSLSVLACRYVDEVIIGSPWEITNDMITTFNISVVVHGTVAEKTLNCELD 1302
Query: 359 PYEVPKSMGMFRLLESPKDITTTTVAQRILANHEAYEKRNAKKSQSEKRYYEEKNYVSGD 418
PYEVPKSMG+F LLESPKDITT TVAQRI+ANHEAY KRNAKK++SE+RYYEEK YVSGD
Sbjct: 1303PYEVPKSMGIFHLLESPKDITTATVAQRIMANHEAYVKRNAKKTRSEQRYYEEKKYVSGD 1482
>TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (61%)
Length = 947
Score = 387 bits (993), Expect = e-108
Identities = 193/287 (67%), Positives = 235/287 (81%), Gaps = 16/287 (5%)
Frame = +2
Query: 146 RLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKAGRYKQIKRTEGVSSTDIVGRIMSSLKD 205
+LF EYKID++IHGDDPC+LPDGTDAYA AKKAGRYKQIKRTEGVSSTDIVGR++ +++
Sbjct: 2 KLFDEYKIDFIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRE 181
Query: 206 QKTCE-DQNGTDVKRQ-------------EESMSKDSHISQFLPTSRRIVQFSNGKGPGP 251
+ E + N + ++RQ + + + IS FLPTSRRIVQFSNG+GPGP
Sbjct: 182 RSIAEKNHNHSSLQRQFSNGHSPKFEAGVSAATASGTRISHFLPTSRRIVQFSNGRGPGP 361
Query: 252 NARIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHERSL 311
++ IVYIDGAFDLFHAGHVEIL+ AR+LGDFLLVGIH+D+TVS RG+H PIM+LHERSL
Sbjct: 362 DSHIVYIDGAFDLFHAGHVEILRLARDLGDFLLVGIHTDQTVSATRGSHRPIMNLHERSL 541
Query: 312 SVLACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKS--LHSERDPYEVPKSMGMF 369
SVLACR+VDEVIIGAPWEI+KDM+TTFNISLVVHGT+AE + E +PY VP SMG+F
Sbjct: 542 SVLACRYVDEVIIGAPWEISKDMLTTFNISLVVHGTIAESNDFKKEECNPYAVPISMGIF 721
Query: 370 RLLESPKDITTTTVAQRILANHEAYEKRNAKKSQSEKRYYEEKNYVS 416
++LESP DITTTT+ +RI+ NHEAY+ RN KK +SEKRYYE K++VS
Sbjct: 722 KVLESPLDITTTTIIRRIVCNHEAYQNRNKKKDESEKRYYEGKSHVS 862
Score = 83.2 bits (204), Expect = 2e-16
Identities = 54/171 (31%), Positives = 88/171 (50%), Gaps = 5/171 (2%)
Frame = +2
Query: 67 VYMDGCFDLMHYGHANALRQAKALGDELVVGLVNDEEILATKGP--PVLTMAERLALVSG 124
VY+DG FDL H GH LR A+ LGD L+VG+ D+ + AT+G P++ + ER V
Sbjct: 374 VYIDGAFDLFHAGHVEILRLARDLGDFLLVGIHTDQTVSATRGSHRPIMNLHERSLSVLA 553
Query: 125 LKWVDEVITDAPYAITEQFLHRLFHEYKIDYVIHG---DDPCLLPDGTDAYAAAKKAGRY 181
++VDEVI AP+ I++ L + I V+HG + + + YA G +
Sbjct: 554 CRYVDEVIIGAPWEISKDML----TTFNISLVVHGTIAESNDFKKEECNPYAVPISMGIF 721
Query: 182 KQIKRTEGVSSTDIVGRIMSSLKDQKTCEDQNGTDVKRQEESMSKDSHISQ 232
K ++ +++T I+ RI+ + + + + KR E SH+S+
Sbjct: 722 KVLESPLDITTTTIIRRIVCNHEAYQNRNKKKDESEKRYYEG---KSHVSE 865
>TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (11%)
Length = 620
Score = 178 bits (452), Expect = 4e-45
Identities = 85/102 (83%), Positives = 96/102 (93%)
Frame = +2
Query: 317 RHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKSLHSERDPYEVPKSMGMFRLLESPK 376
R+VDEVIIG+PWEIT DMITTFNIS+VVHGTV+EKSL+ E DPYE+PKSMG+FRLLESPK
Sbjct: 2 RYVDEVIIGSPWEITNDMITTFNISVVVHGTVSEKSLNCELDPYEIPKSMGIFRLLESPK 181
Query: 377 DITTTTVAQRILANHEAYEKRNAKKSQSEKRYYEEKNYVSGD 418
DITT TVAQRI+ANHEAY KRNAKK++SE+RYYEEK YVSGD
Sbjct: 182 DITTATVAQRIMANHEAYVKRNAKKTRSEQRYYEEKKYVSGD 307
>TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (7%)
Length = 593
Score = 177 bits (450), Expect = 6e-45
Identities = 85/98 (86%), Positives = 90/98 (91%), Gaps = 2/98 (2%)
Frame = +1
Query: 1 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFGGIG--HLPLPFPWSNSGIFHKK 58
MDYE+NSWIWEGV YYP +FGGLMVTAALLGLSTSYFGGIG +L LP WSN GIFH+K
Sbjct: 298 MDYENNSWIWEGVNYYPCVFGGLMVTAALLGLSTSYFGGIGVPYLSLPCSWSNLGIFHQK 477
Query: 59 KSGKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVV 96
KSGKRR+RVYMDGCFDLMHYGHANALRQAKALGDELVV
Sbjct: 478 KSGKRRIRVYMDGCFDLMHYGHANALRQAKALGDELVV 591
Score = 41.6 bits (96), Expect = 7e-04
Identities = 22/52 (42%), Positives = 29/52 (55%)
Frame = +1
Query: 234 LPTSRRIVQFSNGKGPGPNARIVYIDGAFDLFHAGHVEILKRARELGDFLLV 285
LP S + + K G VY+DG FDL H GH L++A+ LGD L+V
Sbjct: 436 LPCSWSNLGIFHQKKSGKRRIRVYMDGCFDLMHYGHANALRQAKALGDELVV 591
>BF009812
Length = 247
Score = 124 bits (311), Expect = 8e-29
Identities = 60/82 (73%), Positives = 70/82 (85%)
Frame = +2
Query: 180 RYKQIKRTEGVSSTDIVGRIMSSLKDQKTCEDQNGTDVKRQEESMSKDSHISQFLPTSRR 239
RYKQIKRTEGVSSTDI GRI+SSL+DQK+C+D NGT+VK QEE+ S+ HI+QF+PT RR
Sbjct: 2 RYKQIKRTEGVSSTDIEGRILSSLRDQKSCDDHNGTEVKPQEENQSQAWHIAQFVPTCRR 181
Query: 240 IVQFSNGKGPGPNARIVYIDGA 261
IVQ NGKGP PN RI Y+DGA
Sbjct: 182 IVQL*NGKGPDPNLRIEYMDGA 247
>TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine
cytidylyltransferase , partial (83%)
Length = 870
Score = 114 bits (286), Expect = 6e-26
Identities = 64/142 (45%), Positives = 85/142 (59%), Gaps = 4/142 (2%)
Frame = +2
Query: 63 RRVRVYMDGCFDLMHYGHANALRQAKAL--GDELVVGLVNDEEILATKGPPVLTMAERLA 120
R VRVY DG +DL H+GHA +L QAK L+VG ND+ KG V+T AER
Sbjct: 92 RPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCNDQVTHKYKGKTVMTEAERYE 271
Query: 121 LVSGLKWVDEVITDAPYAITEQFLHRLFHEYKIDYVIHGDDPCLLPDG--TDAYAAAKKA 178
+ KWVDEVI DAP+ I ++FL + + IDYV H P +G D Y K
Sbjct: 272 SLRHCKWVDEVIPDAPWVINQEFLDK----HNIDYVAHDSLPYADANGAANDVYEFVKSV 439
Query: 179 GRYKQIKRTEGVSSTDIVGRIM 200
GR+K+ KRTEG+S++D++ RI+
Sbjct: 440 GRFKETKRTEGISTSDVIMRIV 505
Score = 88.6 bits (218), Expect = 5e-18
Identities = 55/164 (33%), Positives = 87/164 (52%), Gaps = 7/164 (4%)
Frame = +2
Query: 256 VYIDGAFDLFHAGHVEILKRAREL--GDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSV 313
VY DG +DLFH GH L++A++ +LLVG +D+ +++G +M ER S+
Sbjct: 104 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCNDQVTHKYKGK--TVMTEAERYESL 277
Query: 314 LACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKSLH-SERDPYEVPKSMGMFRLL 372
C+ VDEVI APW I ++ + NI V H ++ + + D YE KS+G F+
Sbjct: 278 RHCKWVDEVIPDAPWVINQEFLDKHNIDYVAHDSLPYADANGAANDVYEFVKSVGRFKET 457
Query: 373 ESPKDITTTTVAQRILANHEAYEKRNAKKSQSEKR----YYEEK 412
+ + I+T+ V RI+ ++ Y RN + S Y +EK
Sbjct: 458 KRTEGISTSDVIMRIVKDYNQYVLRNLDRGYSRNELGVSYVKEK 589
>BE805781
Length = 156
Score = 58.5 bits (140), Expect = 5e-09
Identities = 26/31 (83%), Positives = 30/31 (95%)
Frame = +1
Query: 324 IGAPWEITKDMITTFNISLVVHGTVAEKSLH 354
IG+PWEIT DMITTFNIS+VVHGTVAEK+L+
Sbjct: 1 IGSPWEITNDMITTFNISVVVHGTVAEKTLN 93
>TC229485 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (7%)
Length = 553
Score = 46.2 bits (108), Expect = 3e-05
Identities = 19/29 (65%), Positives = 25/29 (85%)
Frame = +3
Query: 388 LANHEAYEKRNAKKSQSEKRYYEEKNYVS 416
++NHEAY+ RN KK +SEKRYYE K++VS
Sbjct: 3 VSNHEAYQNRNKKKGESEKRYYEGKSHVS 89
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,905,854
Number of Sequences: 63676
Number of extensions: 248293
Number of successful extensions: 1248
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1232
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1237
length of query: 418
length of database: 12,639,632
effective HSP length: 100
effective length of query: 318
effective length of database: 6,272,032
effective search space: 1994506176
effective search space used: 1994506176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0004a.6


