

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.4
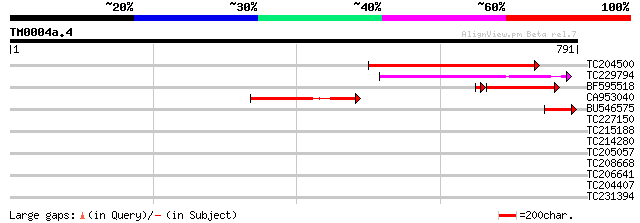
(791 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204500 similar to UP|Q84YG7 (Q84YG7) Isoamylase isoform 1, par... 473 e-133
TC229794 similar to UP|Q9M0S5 (Q9M0S5) Isoamylase-like protein, ... 223 3e-58
BF595518 199 3e-54
CA953040 homologue to PIR|B85091|B85 isoamylase-like protein [im... 167 2e-41
BU546575 91 3e-18
TC227150 similar to UP|Q9ARE4 (Q9ARE4) ZF-HD homeobox protein, p... 31 2.4
TC215188 homologue to UP|Q9XIS5 (Q9XIS5) Starch branching enzyme... 30 5.4
TC214280 UP|Q93YG3 (Q93YG3) Chlorophyll a/b binding protein type... 30 5.4
TC205057 homologue to UP|Q43448 (Q43448) Cysteine proteinase pre... 29 9.2
TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein D... 29 9.2
TC206641 UP|Q43448 (Q43448) Cysteine proteinase precursor , part... 29 9.2
TC204407 similar to UP|O48618 (O48618) Cytochome b5 (Fragment), ... 29 9.2
TC231394 similar to UP|Q9LW11 (Q9LW11) Zinc finger protein-like;... 29 9.2
>TC204500 similar to UP|Q84YG7 (Q84YG7) Isoamylase isoform 1, partial (30%)
Length = 719
Score = 473 bits (1218), Expect = e-133
Identities = 217/239 (90%), Positives = 225/239 (93%)
Frame = +2
Query: 501 DTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKH 560
DTVR F+KGTDGFAGAFAECLCGSPN+YQGGGRKPW+SINFVC HDGFTLADLVTYNNK+
Sbjct: 2 DTVRLFIKGTDGFAGAFAECLCGSPNLYQGGGRKPWHSINFVCAHDGFTLADLVTYNNKN 181
Query: 561 NLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDE 620
NL NGEDNNDGENHN SWNCGQEGEF S+SVKKLRKRQMRNFFLSLMVSQGVPMIYMGDE
Sbjct: 182 NLSNGEDNNDGENHNYSWNCGQEGEFVSTSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDE 361
Query: 621 YGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRFCCLMTKFRYECESLGLDDFPTSDR 680
YGHTKGGNNNTYCHDNY NYFQWD KEESSSDFFRFC LMTKFR ECESLGL DFPTS+R
Sbjct: 362 YGHTKGGNNNTYCHDNYHNYFQWDKKEESSSDFFRFCRLMTKFRQECESLGLADFPTSER 541
Query: 681 LQWHGHFPGKPDWSETSRFVAFTMVDSVKREIYIAFNTSHLPVTITLPERPGYRWEPLV 739
LQWHGHFPGKPDWSETSRFVA TMVDSVK EIYIAFN SHLP T+TLPERPGY+WEPLV
Sbjct: 542 LQWHGHFPGKPDWSETSRFVACTMVDSVKGEIYIAFNMSHLPFTVTLPERPGYKWEPLV 718
>TC229794 similar to UP|Q9M0S5 (Q9M0S5) Isoamylase-like protein, partial
(36%)
Length = 1141
Score = 223 bits (567), Expect = 3e-58
Identities = 118/269 (43%), Positives = 159/269 (58%), Gaps = 1/269 (0%)
Frame = +2
Query: 516 AFAECLCGSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHN 575
+ A + GS ++Y+ R+P++SINFV HDGFTL DLV+YN KHN NGE DG N N
Sbjct: 11 SLATRVAGSSDLYRVNNRRPYHSINFVIAHDGFTLRDLVSYNFKHNKANGEGGKDGSNDN 190
Query: 576 NSWNCGQEGEFASSSVKKLRKRQMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHD 635
SWNCG EGE +S++ LR RQM+NF L+LM+SQG PM+ MGDEYGHT+ GNNN+Y HD
Sbjct: 191 FSWNCGFEGETDDASIRALRSRQMKNFHLALMISQGTPMMLMGDEYGHTRYGNNNSYGHD 370
Query: 636 NYLNYFQWDIKEESSSDFFRFCCLMTKFRYECESLGLDDFPTSDRLQWHGHFPGKPDWSE 695
+N F WD + SD FRF + K+R+ E ++F ++ + WH PD
Sbjct: 371 TAINNFLWDQLDARKSDHFRFFSKVIKYRHAHEVFSRENFLNTNDITWHEDNWENPD--- 541
Query: 696 TSRFVAFTMVDSVKREIYIAFNTSHLPVTITLPERPGYR-WEPLVDTGKSAPYDFLTPDL 754
S+F+AFT+ D +IY+AFN+ V + LP P R W + DT +P DF+ +
Sbjct: 542 -SKFLAFTLHDRSGGDIYVAFNSHDYFVKVLLPTPPKKRNWFRVADTNLKSPDDFVLDGV 718
Query: 755 PGRDIAIQQYAQFLDANLYPMLSYSSIIL 783
P N Y + YSSI+L
Sbjct: 719 PS------------VGNTYNIAPYSSILL 769
>BF595518
Length = 422
Score = 199 bits (505), Expect(2) = 3e-54
Identities = 91/102 (89%), Positives = 94/102 (91%)
Frame = +3
Query: 666 ECESLGLDDFPTSDRLQWHGHFPGKPDWSETSRFVAFTMVDSVKREIYIAFNTSHLPVTI 725
ECESLGL DFPTS+RLQWHGHFPGKPDWSETSRFVA TMVDSVK EIYIAFN SHLP T+
Sbjct: 117 ECESLGLADFPTSERLQWHGHFPGKPDWSETSRFVACTMVDSVKGEIYIAFNMSHLPFTV 296
Query: 726 TLPERPGYRWEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQF 767
TLPERPGY+WEPLVDT K PYDFLTPDLPGRDIAIQQYAQF
Sbjct: 297 TLPERPGYKWEPLVDTSKPTPYDFLTPDLPGRDIAIQQYAQF 422
Score = 32.7 bits (73), Expect(2) = 3e-54
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = +2
Query: 650 SSDFFRFCCLMTKFR 664
SSDFFRFC LMTKFR
Sbjct: 2 SSDFFRFCRLMTKFR 46
>CA953040 homologue to PIR|B85091|B85 isoamylase-like protein [imported] -
Arabidopsis thaliana, partial (19%)
Length = 421
Score = 167 bits (422), Expect = 2e-41
Identities = 81/154 (52%), Positives = 106/154 (68%), Gaps = 1/154 (0%)
Frame = +1
Query: 337 IEVIMDVVFNHTVEGNENGPII-SFRGVDNSIYYMIAPKGEFYNYSGCGNTLNCNHPVVR 395
++VI+DVV+NHT E ++ P SFRG+DN +YYM+ G+ N+SGCGNTLNCNHPVV
Sbjct: 10 MQVILDVVYNHTNEADDAFPYTTSFRGIDNKVYYMLDNNGQLLNFSGCGNTLNCNHPVVM 189
Query: 396 QFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPL 455
+ I+D LR+WVTE HVDGFRFDLAS++ R G++ G+PL +PPL
Sbjct: 190 ELILDSLRHWVTEYHVDGFRFDLASVLCR------GID-------------GSPLNAPPL 312
Query: 456 IDLISNDPILHGVKLIAEAWDAGGLYQVGTFPHW 489
I I+ D +L K+IAE WD GGLY VG+FP+W
Sbjct: 313 IRAIAKDAVLSRCKIIAEPWDCGGLYLVGSFPNW 414
>BU546575
Length = 457
Score = 90.5 bits (223), Expect = 3e-18
Identities = 42/45 (93%), Positives = 43/45 (95%)
Frame = -2
Query: 746 PYDFLTPDLPGRDIAIQQYAQFLDANLYPMLSYSSIILLRTPDEN 790
PYDFLTPD PGRDIAIQQYAQFLDAN+YPMLSYSSIILLR PDEN
Sbjct: 456 PYDFLTPDXPGRDIAIQQYAQFLDANMYPMLSYSSIILLRIPDEN 322
>TC227150 similar to UP|Q9ARE4 (Q9ARE4) ZF-HD homeobox protein, partial (32%)
Length = 969
Score = 30.8 bits (68), Expect = 2.4
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Frame = +3
Query: 556 YNNKHNLPNGEDNN--DGENHNNSWNCGQEGEFASSSVKKLRKR 597
+NNKH L N E ++ +G NH NS + +F+ + K R+R
Sbjct: 435 HNNKHTLANSEQHSAGNGSNHLNSTTIIENSQFS*GILTK*RER 566
>TC215188 homologue to UP|Q9XIS5 (Q9XIS5) Starch branching enzyme , partial
(48%)
Length = 1878
Score = 29.6 bits (65), Expect = 5.4
Identities = 21/96 (21%), Positives = 42/96 (42%)
Frame = +3
Query: 373 PKGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGV 432
P Y++ N V ++++ R+W+ E DGFRFD + M + +G+
Sbjct: 12 PGSRGYHWMWDSRLFNYGSWEVLRYLLSNARWWLDEYKFDGFRFDGVTSMMYTH---HGL 182
Query: 433 NVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGV 468
V T + T + ++ L+ + ++HG+
Sbjct: 183 EVAFTGNYNEYFGFATDV--DAVVYLMLTNDVIHGL 284
>TC214280 UP|Q93YG3 (Q93YG3) Chlorophyll a/b binding protein type II
precursor, partial (38%)
Length = 1174
Score = 29.6 bits (65), Expect = 5.4
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 8/50 (16%)
Frame = +3
Query: 262 LGVNCIELL--------PCHEFNELEYFSYNSVQGDYRVNFWGYSTVNYF 303
L N I+LL P +FN L YF Y + + + FW +ST+ F
Sbjct: 450 LNFNFIKLLLFTIFLSSPLGQFNALAYFPYYFTRPNQMLPFWWHSTIYVF 599
>TC205057 homologue to UP|Q43448 (Q43448) Cysteine proteinase precursor ,
complete
Length = 1457
Score = 28.9 bits (63), Expect = 9.2
Identities = 22/82 (26%), Positives = 36/82 (43%)
Frame = +2
Query: 418 LASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDA 477
+A+ + ++ L GVN + T VS PLI S + HGV L+
Sbjct: 866 IAAYLVKNGPLAMGVNAI-------FMQTYIGGVSCPLI--CSKKRLNHGVLLVGYGAKG 1018
Query: 478 GGLYQVGTFPHWGIWSEWNGKY 499
+ ++G P+W I + W K+
Sbjct: 1019FSILRLGNKPYWIIKNSWGEKW 1084
>TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein DG1037
(Fragment), partial (4%)
Length = 1021
Score = 28.9 bits (63), Expect = 9.2
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = -2
Query: 555 TYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRKRQM 599
T NN +N N +NN+ N+NN+ N + S KK+ +M
Sbjct: 285 TCNNNNNNNNNNNNNNNNNNNNNNNIKLHKKTGSKLNKKIAPIKM 151
>TC206641 UP|Q43448 (Q43448) Cysteine proteinase precursor , partial (95%)
Length = 1407
Score = 28.9 bits (63), Expect = 9.2
Identities = 22/82 (26%), Positives = 36/82 (43%)
Frame = +2
Query: 418 LASIMTRSSSLWNGVNVFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDA 477
+A+ + ++ L GVN + T VS PLI S + HGV L+
Sbjct: 785 IAAYLVKNGPLAMGVNAI-------FMQTYIGGVSCPLI--CSKKRLNHGVLLVGYGAKG 937
Query: 478 GGLYQVGTFPHWGIWSEWNGKY 499
+ ++G P+W I + W K+
Sbjct: 938 FSILRLGNKPYWIIKNSWGEKW 1003
>TC204407 similar to UP|O48618 (O48618) Cytochome b5 (Fragment), partial
(78%)
Length = 857
Score = 28.9 bits (63), Expect = 9.2
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 5/76 (6%)
Frame = +3
Query: 709 KREIYIAFNTSHLPVTITLPERPGYRWEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQFL 768
K++ +I N +T L E PG L TGK A DF D+ D AI+ ++
Sbjct: 201 KKDCWIIINGKVYDITPFLDEHPGGDEVLLTSTGKDATIDF--EDVGHSDSAIEMMEKYF 374
Query: 769 -----DANLYPMLSYS 779
+ L P +S+S
Sbjct: 375 IGKVDTSTLPPKVSHS 422
>TC231394 similar to UP|Q9LW11 (Q9LW11) Zinc finger protein-like; Ser/Thr
protein kinase-like protein, partial (62%)
Length = 689
Score = 28.9 bits (63), Expect = 9.2
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 8/41 (19%)
Frame = +2
Query: 515 GAFAECLCGSPN--------VYQGGGRKPWNSINFVCTHDG 547
GAF + L GS N + G GR W S +++CT G
Sbjct: 215 GAFKDDLAGSYNSSDILRSRAFGGSGRPGWKSGDWICTRSG 337
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,443,573
Number of Sequences: 63676
Number of extensions: 641952
Number of successful extensions: 2981
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 2862
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2923
length of query: 791
length of database: 12,639,632
effective HSP length: 105
effective length of query: 686
effective length of database: 5,953,652
effective search space: 4084205272
effective search space used: 4084205272
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0004a.4


