Nucleotide Sequence (with vector) for pFN21AB9849
Download
>pFN21AB9849 5836 bp
TCAATATTGGCCATTAGCCATATTATTCATTGGTTATATAGCATAAATCAATATTGGCTA
TTGGCCATTGCATACGTTGTATCTATATCATAATATGTACATTTATATTGGCTCATGTCC
AATATGACCGCCATGTTGGCATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGG
GTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCC
GCCTGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT
AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGC
CCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTCCGCCCCCTATTGACGTCAATGA
CGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTACGGGACTTTCCTACTTG
GCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACAC
CAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGT
CAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAATAACCC
CGCCCCGTTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGC
TGGTTTAGTGAACCGTCAGATCACTAGAAGCTTTATTGCGGTAGTTTATCACAGTTAAAT
TGCTAACGCAGTCAGTGCTTCTGACACAACAGTCTCGAACTTAAGCTGCAGAAGTTGGTC
GTGAGGCACTGGGCAGGTAAGTATCAAGGTTACAAGACAGGTTTAAGGAGACCAATAGAA
ACTGGGCTTGTCGAGACAGAGAAGACTCTTGCGTTTCTGATAGGCACCTATTGGTCTTAC
TGACATCCACTTTGCCTTTCTCTCCACAGGTGTCCACTCCCAGTTCAATTACAGCTCTTA
AGGCTAGAGTATTAATACGACTCACTATAGGGCTAGCAAAGCCACCATGGCAGAAATCGG
TACTGGCTTTCCATTCGACCCCCATTATGTGGAAGTCCTGGGCGAGCGCATGCACTACGT
CGATGTTGGTCCGCGCGATGGCACCCCTGTGCTGTTCCTGCACGGTAACCCGACCTCCTC
CTACGTGTGGCGCAACATCATCCCGCATGTTGCACCGACCCATCGCTGCATTGCTCCAGA
CCTGATCGGTATGGGCAAATCCGACAAACCAGACCTGGGTTATTTCTTCGACGACCACGT
CCGCTTCATGGATGCCTTCATCGAAGCCCTGGGTCTGGAAGAGGTCGTCCTGGTCATTCA
CGACTGGGGCTCCGCTCTGGGTTTCCACTGGGCCAAGCGCAATCCAGAGCGCGTCAAAGG
TATTGCATTTATGGAGTTCATCCGCCCTATCCCGACCTGGGACGAATGGCCAGAATTTGC
CCGCGAGACCTTCCAGGCCTTCCGCACCACCGACGTCGGCCGCAAGCTGATCATCGATCA
GAACGTTTTTATCGAGGGTACGCTGCCGATGGGTGTCGTCCGCCCGCTGACTGAAGTCGA
GATGGACCATTACCGCGAGCCGTTCCTGAATCCTGTTGACCGCGAGCCACTGTGGCGCTT
CCCAAACGAGCTGCCAATCGCCGGTGAGCCAGCGAACATCGTCGCGCTGGTCGAAGAATA
CATGGACTGGCTGCACCAGTCCCCTGTCCCGAAGCTGCTGTTCTGGGGCACCCCAGGCGT
TCTGATCCCACCGGCCGAAGCCGCTCGCCTGGCCAAAAGCCTGCCTAACTGCAAGGCTGT
GGACATCGGCCCGGGTCTGAATCTGCTGCAAGAAGACAACCCGGACCTGATCGGCAGCGA
GATCGCGCGCTGGCTGTCGACGCTCGAGATTTCCGGCGAGCCAACCACTGAGGATCTGTA
CTTTCAGAGCGATAACGCGATCGCCATGGCAGCAGACCTGGGCCCCTGGAATGACACCAT
CAATGGCACCTGGGATGGGGATGAGCTGGGCTACAGGTGCCGCTTCAACGAGGACTTCAA
GTACGTGCTGCTGCCTGTGTCCTACGGCGTGGTGTGCGTGCTTGGGCTGTGTCTGAACGC
CGTGGCGCTCTACATCTTCTTGTGCCGCCTCAAGACCTGGAATGCGTCCACCACATATAT
GTTCCACCTGGCTGTGTCTGATGCACTGTATGCGGCCTCCCTGCCGCTGCTGGTCTATTA
CTACGCCCGCGGCGACCACTGGCCCTTCAGCACGGTGCTCTGCAAGCTGGTGCGCTTCCT
CTTCTACACCAACCTTTACTGCAGCATCCTCTTCCTCACCTGCATCAGCGTGCACCGGTG
TCTGGGCGTCTTACGACCTCTGCGCTCCCTGCGCTGGGGCCGGGCCCGCTACGCTCGCCG
GGTGGCCGGGGCCGTGTGGGTGTTGGTGCTGGCCTGCCAGGCCCCCGTGCTCTACTTTGT
CACCACCAGCGCGCGCGGGGGCCGCGTAACCTGCCACGACACCTCGGCACCCGAGCTCTT
CAGCCGCTTCGTGGCCTACAGCTCAGTCATGCTGGGCCTGCTCTTCGCGGTGCCCTTTGC
CGTCATCCTTGTCTGTTACGTGCTCATGGCTCGGCGACTGCTAAAGCCAGCCTACGGGAC
CTCGGGCGGCCTGCCTAGGGCCAAGCGCAAGTCCGTGCGCACCATCGCCGTGGTGCTGGC
TGTCTTCGCCCTCTGCTTCCTGCCATTCCACGTCACCCGCACCCTCTACTACTCCTTCCG
TTCGCTGGACCTCAGCTGCCACACCCTCAACGCCATCAACATGGCCTACAAGGTTACCCG
GCCGCTGGCCAGTGCTAACAGTTGCCTTGACCCCGTGCTCTACTTCCTGGCTGGGCAGAG
GCTCGTACGCTTTGCCCGAGATGCCAAGCCACCCACTGGCCCCAGCCCTGCCACCCCGGC
TCGCTGCAGGCTGGGCCTGCGCAGATCCGACAGAACTGACATGCAGAGGATAGAAGATGT
GTTGGGCAGCAGTGAGGACTCTAGGCGGACAGAGTCCACGCCGGCTGGTAGCGAGAACAC
TAAGGACATTCGGCTGGTTTAAACGAATTCGGGCTCGGTACCCGGGGATCCTCTAGAGTC
GACCTGCAGGCATGCAAGCTGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGTTGG
CTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCGGCCGCTTCGAGCAGAC
ATGATAAGATACATTGATGAGTTTGGACAAACCACAACTAGAATGCAGTGAAAAAAATGC
TTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAACCATTATAAGCTGCAATAAA
CAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGGGAGATGTGGGAG
GTTTTTTTAAGCAAGTAAAACCTCTACAAATGTGGTAAAATCGAATTCTAATGGATCCTC
TTTGCGCTTGCGTTTTCCCTTGTCCAGATAGCCCAGTAGCTGACATTCATCCGGGGTCAG
CACCGTTTCTGCGGACTGGCTTTCTACGTAATGGTTTCTTAGACGTCAGGTGGCACTTTT
CGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATACATTCAAATATGTAT
CCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAGTATG
AGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCTTCCTGTT
TTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCACGA
GTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCGCCCCGAA
GAACGTTTTCCAATGATGAGCACTTTCAAAGTTCTGCTATGTGGCGCGGTATTATCCCGT
ATTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACTTGGTT
GAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGC
AGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAACTATCGGA
GGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCGCCTTGAT
CGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCT
GTAGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGCTTCC
CGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCG
GCCCTTCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGC
GGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTATCTACACG
ACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCA
CTGATTAAGCATTGGTAATTCGAAATGACCGACCAAGCGACGCCCAACCGGTATCAGCTC
ACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGT
GAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCC
ATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAA
ACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTC
CTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGG
CGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGC
TGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATC
GTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACA
GGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACT
ACGGCTACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCG
GAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTT
TTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATTTCAAGAAGATCCTTTGATCT
TTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGA
GATTATCAAAAAGGATCTTCACCTAGATCCTTTTATAGTCCGGAAATACAGGAACGCACG
CTGGATGGCCCTTCGCTGGGATGGTGAAACCATGAAAAATGGCAGCTTCAGTGGATTAAG
TGGGGGTAATGTGGCCTGTACCCTCTGGTTGCATAGGTATTCATACGGTTAAAATTTATC
AGGCGCGATTGCGGCAGTTTTTCGGGTGGTTTGTTGCCATTTTTACCTGTCTGCTGCCGT
GATCGCGCTGAACGCGTTTTAGCGGTGCGTACAATTAAGGGATTATGGTAAATCCACTTA
CTGTCTGCCCTCGTAGCCATCGAGATAAACCGCAGTACTCCGGCCACGATGCGTCCGGCG
TAGAGGATCGAGATCT
 Characterization of the cloned ORF
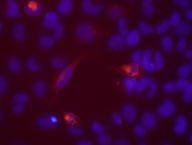
Characterization of the cloned ORF N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand
N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand Length: 1131 bp
Length: 1131 bp Restriction map A
Restriction map A
 Restriction map B
Restriction map B Genomic Structure
Genomic Structure Length: 377 aa
Length: 377 aa
 Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)
Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)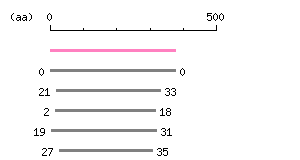
 Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]
Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]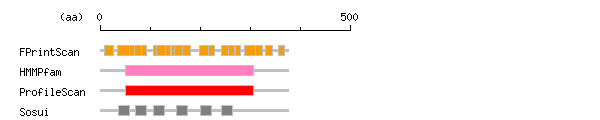
 Result of InterProScan
Result of InterProScan Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)
Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)


 more Linker info
more Linker info