Nucleotide Sequence (with vector) for pFN21AB8810
Download
>pFN21AB8810 5788 bp
TCAATATTGGCCATTAGCCATATTATTCATTGGTTATATAGCATAAATCAATATTGGCTA
TTGGCCATTGCATACGTTGTATCTATATCATAATATGTACATTTATATTGGCTCATGTCC
AATATGACCGCCATGTTGGCATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGG
GTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCC
GCCTGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT
AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGC
CCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTCCGCCCCCTATTGACGTCAATGA
CGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTACGGGACTTTCCTACTTG
GCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACAC
CAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGT
CAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAATAACCC
CGCCCCGTTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGC
TGGTTTAGTGAACCGTCAGATCACTAGAAGCTTTATTGCGGTAGTTTATCACAGTTAAAT
TGCTAACGCAGTCAGTGCTTCTGACACAACAGTCTCGAACTTAAGCTGCAGAAGTTGGTC
GTGAGGCACTGGGCAGGTAAGTATCAAGGTTACAAGACAGGTTTAAGGAGACCAATAGAA
ACTGGGCTTGTCGAGACAGAGAAGACTCTTGCGTTTCTGATAGGCACCTATTGGTCTTAC
TGACATCCACTTTGCCTTTCTCTCCACAGGTGTCCACTCCCAGTTCAATTACAGCTCTTA
AGGCTAGAGTATTAATACGACTCACTATAGGGCTAGCAAAGCCACCATGGCAGAAATCGG
TACTGGCTTTCCATTCGACCCCCATTATGTGGAAGTCCTGGGCGAGCGCATGCACTACGT
CGATGTTGGTCCGCGCGATGGCACCCCTGTGCTGTTCCTGCACGGTAACCCGACCTCCTC
CTACGTGTGGCGCAACATCATCCCGCATGTTGCACCGACCCATCGCTGCATTGCTCCAGA
CCTGATCGGTATGGGCAAATCCGACAAACCAGACCTGGGTTATTTCTTCGACGACCACGT
CCGCTTCATGGATGCCTTCATCGAAGCCCTGGGTCTGGAAGAGGTCGTCCTGGTCATTCA
CGACTGGGGCTCCGCTCTGGGTTTCCACTGGGCCAAGCGCAATCCAGAGCGCGTCAAAGG
TATTGCATTTATGGAGTTCATCCGCCCTATCCCGACCTGGGACGAATGGCCAGAATTTGC
CCGCGAGACCTTCCAGGCCTTCCGCACCACCGACGTCGGCCGCAAGCTGATCATCGATCA
GAACGTTTTTATCGAGGGTACGCTGCCGATGGGTGTCGTCCGCCCGCTGACTGAAGTCGA
GATGGACCATTACCGCGAGCCGTTCCTGAATCCTGTTGACCGCGAGCCACTGTGGCGCTT
CCCAAACGAGCTGCCAATCGCCGGTGAGCCAGCGAACATCGTCGCGCTGGTCGAAGAATA
CATGGACTGGCTGCACCAGTCCCCTGTCCCGAAGCTGCTGTTCTGGGGCACCCCAGGCGT
TCTGATCCCACCGGCCGAAGCCGCTCGCCTGGCCAAAAGCCTGCCTAACTGCAAGGCTGT
GGACATCGGCCCGGGTCTGAATCTGCTGCAAGAAGACAACCCGGACCTGATCGGCAGCGA
GATCGCGCGCTGGCTGTCGACGCTCGAGATTTCCGGCGAGCCAACCACTGAGGATCTGTA
CTTTCAGAGCGATAACGCGATCGCCATGGATATACAAATGGCAAACAATTTTACTCCGCC
CTCTGCAACTCCTCAGGGAAATGACTGTGACCTCTATGCACATCACAGCACGGCCAGGAT
AGTAATGCCTCTGCATTACAGCCTCGTCTTCATCATTGGGCTCGTGGGAAACTTACTAGC
CTTGGTCGTCATTGTTCAAAACAGGAAAAAAATCAACTCTACCACCCTCTATTCAACAAA
TTTGGTGATTTCTGATATACTTTTTACCACCGCTTTGCCTACACGAATAGCCTACTATGC
AATGGGCTTTGACTGGAGAATCGGAGATGCCTTGTGTAGGATAACTGCGCTAGTGTTTTA
CATCAACACATATGCAGGTGTGAACTTTATGACCTGCCTGAGTATTGACCGCTTCATTGC
TGTGGTGCACCCTCTACGCTACAACAAGATAAAAAGGATTGAACATGCAAAAGGCGTGTG
CATATTTGTCTGGATTCTAGTATTTGCTCAGACACTCCCACTCCTCATCAACCCTATGTC
AAAGCAGGAGGCTGAAAGGATTACATGCATGGAGTATCCAAACTTTGAAGAAACTAAATC
TCTTCCCTGGATTCTGCTTGGGGCATGTTTCATAGGATATGTACTTCCACTTATAATCAT
TCTCATCTGCTATTCTCAGATCTGCTGCAAACTCTTCAGAACTGCCAAACAAAACCCACT
CACTGAGAAATCTGGTGTAAACAAAAAGGCTCTCAACACAATTATTCTTATTATTGTTGT
GTTTGTTCTCTGTTTCACACCTTACCATGTTGCAATTATTCAACATATGATTAAGAAGCT
TCGTTTCTCTAATTTCCTGGAATGTAGCCAAAGACATTCGTTCCAGATTTCTCTGCACTT
TACAGTATGCCTGATGAACTTCAATTGCTGCATGGACCCTTTTATCTACTTCTTTGCATG
TAAAGGGTATAAGAGAAAGGTTATGAGGATGCTGAAACGGCAAGTCAGTGTATCGATTTC
TAGTGCTGTGAAGTCAGCCCCTGAAGAAAATTCACGTGAAATGACAGAAACGCAGATGAT
GATACATTCCAAGTCTTCAAATGGAAAGGTTTAAACGAATTCGGGCTCGGTACCCGGGGA
TCCTCTAGAGTCGACCTGCAGGCATGCAAGCTGATCCGGCTGCTAACAAAGCCCGAAAGG
AAGCTGAGTTGGCTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCGGCCG
CTTCGAGCAGACATGATAAGATACATTGATGAGTTTGGACAAACCACAACTAGAATGCAG
TGAAAAAAATGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAACCATTATA
AGCTGCAATAAACAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGG
GAGATGTGGGAGGTTTTTTTAAGCAAGTAAAACCTCTACAAATGTGGTAAAATCGAATTC
TAATGGATCCTCTTTGCGCTTGCGTTTTCCCTTGTCCAGATAGCCCAGTAGCTGACATTC
ATCCGGGGTCAGCACCGTTTCTGCGGACTGGCTTTCTACGTAATGGTTTCTTAGACGTCA
GGTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATACAT
TCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAA
AGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTT
TGCCTTCCTGTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAG
TTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGT
TTTCGCCCCGAAGAACGTTTTCCAATGATGAGCACTTTCAAAGTTCTGCTATGTGGCGCG
GTATTATCCCGTATTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAG
AATGACTTGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTA
AGAGAATTATGCAGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTG
ACAACTATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTA
ACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGAC
ACCACGATGCCTGTAGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTT
ACTCTAGCTTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCA
CTTCTGCGCTCGGCCCTTCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAG
CGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTA
GTTATCTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAG
ATAGGTGCCTCACTGATTAAGCATTGGTAATTCGAAATGACCGACCAAGCGACGCCCAAC
CGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAG
GAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGC
TGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTC
AGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCC
TCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTT
CGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCG
TTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTAT
CCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAG
CCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGT
GGTGGCCTAACTACGGCTACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGC
CAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTA
GCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATTTCAAGAAG
ATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGA
TTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTATAGTCCGGAAATA
CAGGAACGCACGCTGGATGGCCCTTCGCTGGGATGGTGAAACCATGAAAAATGGCAGCTT
CAGTGGATTAAGTGGGGGTAATGTGGCCTGTACCCTCTGGTTGCATAGGTATTCATACGG
TTAAAATTTATCAGGCGCGATTGCGGCAGTTTTTCGGGTGGTTTGTTGCCATTTTTACCT
GTCTGCTGCCGTGATCGCGCTGAACGCGTTTTAGCGGTGCGTACAATTAAGGGATTATGG
TAAATCCACTTACTGTCTGCCCTCGTAGCCATCGAGATAAACCGCAGTACTCCGGCCACG
ATGCGTCCGGCGTAGAGGATCGAGATCT
 Characterization of the cloned ORF
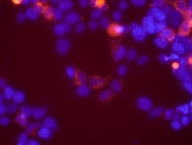
Characterization of the cloned ORF N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand
N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand Length: 1083 bp
Length: 1083 bp Restriction map A
Restriction map A
 Restriction map B
Restriction map B Genomic Structure
Genomic Structure Length: 361 aa
Length: 361 aa
 Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)
Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)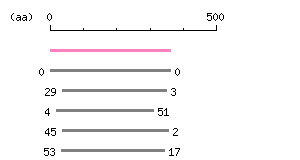
 Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]
Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]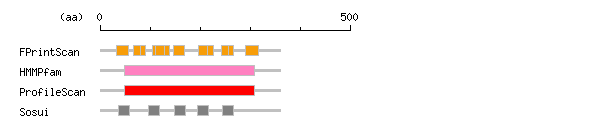
 Result of InterProScan
Result of InterProScan Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)
Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)


 more Linker info
more Linker info