Nucleotide Sequence (with vector) for pFN21AA0001
Download
>pFN21AA0001 5719 bp
TCAATATTGGCCATTAGCCATATTATTCATTGGTTATATAGCATAAATCAATATTGGCTA
TTGGCCATTGCATACGTTGTATCTATATCATAATATGTACATTTATATTGGCTCATGTCC
AATATGACCGCCATGTTGGCATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGG
GTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCC
GCCTGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT
AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGC
CCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTCCGCCCCCTATTGACGTCAATGA
CGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTACGGGACTTTCCTACTTG
GCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACAC
CAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGT
CAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAATAACCC
CGCCCCGTTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGC
TGGTTTAGTGAACCGTCAGATCACTAGAAGCTTTATTGCGGTAGTTTATCACAGTTAAAT
TGCTAACGCAGTCAGTGCTTCTGACACAACAGTCTCGAACTTAAGCTGCAGAAGTTGGTC
GTGAGGCACTGGGCAGGTAAGTATCAAGGTTACAAGACAGGTTTAAGGAGACCAATAGAA
ACTGGGCTTGTCGAGACAGAGAAGACTCTTGCGTTTCTGATAGGCACCTATTGGTCTTAC
TGACATCCACTTTGCCTTTCTCTCCACAGGTGTCCACTCCCAGTTCAATTACAGCTCTTA
AGGCTAGAGTATTAATACGACTCACTATAGGGCTAGCAAAGCCACCATGGCAGAAATCGG
TACTGGCTTTCCATTCGACCCCCATTATGTGGAAGTCCTGGGCGAGCGCATGCACTACGT
CGATGTTGGTCCGCGCGATGGCACCCCTGTGCTGTTCCTGCACGGTAACCCGACCTCCTC
CTACGTGTGGCGCAACATCATCCCGCATGTTGCACCGACCCATCGCTGCATTGCTCCAGA
CCTGATCGGTATGGGCAAATCCGACAAACCAGACCTGGGTTATTTCTTCGACGACCACGT
CCGCTTCATGGATGCCTTCATCGAAGCCCTGGGTCTGGAAGAGGTCGTCCTGGTCATTCA
CGACTGGGGCTCCGCTCTGGGTTTCCACTGGGCCAAGCGCAATCCAGAGCGCGTCAAAGG
TATTGCATTTATGGAGTTCATCCGCCCTATCCCGACCTGGGACGAATGGCCAGAATTTGC
CCGCGAGACCTTCCAGGCCTTCCGCACCACCGACGTCGGCCGCAAGCTGATCATCGATCA
GAACGTTTTTATCGAGGGTACGCTGCCGATGGGTGTCGTCCGCCCGCTGACTGAAGTCGA
GATGGACCATTACCGCGAGCCGTTCCTGAATCCTGTTGACCGCGAGCCACTGTGGCGCTT
CCCAAACGAGCTGCCAATCGCCGGTGAGCCAGCGAACATCGTCGCGCTGGTCGAAGAATA
CATGGACTGGCTGCACCAGTCCCCTGTCCCGAAGCTGCTGTTCTGGGGCACCCCAGGCGT
TCTGATCCCACCGGCCGAAGCCGCTCGCCTGGCCAAAAGCCTGCCTAACTGCAAGGCTGT
GGACATCGGCCCGGGTCTGAATCTGCTGCAAGAAGACAACCCGGACCTGATCGGCAGCGA
GATCGCGCGCTGGCTGTCGACGCTCGAGATTTCCGGCGAGCCAACCACTGAGGATCTGTA
CTTTCAGAGCGATAACGCGATCGCCATGATCAATTCAACCTCCACACAGCCTCCAGATGA
ATCCTGCTCTCAGAACCTCCTGATCACTCAGCAGATCATTCCTGTGCTGTACTGTATGGT
CTTCATTGCAGGAATCCTACTCAATGGAGTGTCAGGATGGATATTCTTTTACGTGCCCAG
CTCTAAGAGTTTCATCATCTATCTCAAGAACATTGTTATTGCTGACTTTGTGATGAGCCT
GACTTTTCCTTTCAAGATCCTTGGTGACTCAGGCCTTGGTCCCTGGCAGCTGAACGTGTT
TGTGTGCAGGGTCTCTGCCGTGCTCTTCTACGTCAACATGTACGTCAGCATTGTGTTCTT
TGGGCTCATCAGCTTTGACAGATATTATAAAATTGTAAAGCCTCTTTGGACTTCTTTCAT
CCAGTCAGTGAGTTACAGCAAACTTCTGTCAGTGATAGTATGGATGCTCATGCTCCTCCT
TGCTGTTCCAAATATTATTCTCACCAACCAGAGTGTTAGGGAGGTTACACAAATAAAATG
TATAGAACTGAAAAGTGAACTGGGACGGAAGTGGCACAAAGCATCAAACTACATCTTCGT
GGCCATCTTCTGGATTGTGTTTCTTTTGTTAATCGTTTTCTATACTGCTATCACAAAGAA
AATCTTTAAGTCCCACCTTAAGTCAAGTCGGAATTCCACTTCGGTCAAAAAGAAATCTAG
CCGCAACATATTCAGCATCGTGTTTGTGTTTTTTGTCTGTTTTGTACCTTACCATATTGC
CAGAATCCCCTACACAAAGAGTCAGACCGAAGCTCATTACAGCTGCCAGTCAAAAGAAAT
CTTGCGGTATATGAAAGAATTCACTCTGCTACTATCTGCTGCAAATGTATGCTTGGACCC
TATTATTTATTTCTTTCTATGCCAGCCGTTTAGGGAAATCTTATGTAAGAAATTGCACAT
TCCATTAAAAGCTCAGAATGACCTAGACATTTCCAGAATCAAAAGAGGAAATACAACACT
TGAAAGCACAGATACTTTGGTTTAAACGAATTCGGGCTCGGTACCCGGGGATCCTCTAGA
GTCGACCTGCAGGCATGCAAGCTGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGT
TGGCTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCGGCCGCTTCGAGCA
GACATGATAAGATACATTGATGAGTTTGGACAAACCACAACTAGAATGCAGTGAAAAAAA
TGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAACCATTATAAGCTGCAAT
AAACAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGGGAGATGTGG
GAGGTTTTTTTAAGCAAGTAAAACCTCTACAAATGTGGTAAAATCGAATTCTAATGGATC
CTCTTTGCGCTTGCGTTTTCCCTTGTCCAGATAGCCCAGTAGCTGACATTCATCCGGGGT
CAGCACCGTTTCTGCGGACTGGCTTTCTACGTAATGGTTTCTTAGACGTCAGGTGGCACT
TTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATACATTCAAATATG
TATCCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAGT
ATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCTTCCT
GTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCA
CGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCGCCCC
GAAGAACGTTTTCCAATGATGAGCACTTTCAAAGTTCTGCTATGTGGCGCGGTATTATCC
CGTATTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACTTG
GTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTA
TGCAGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAACTATC
GGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCGCCTT
GATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATG
CCTGTAGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGCT
TCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGC
TCGGCCCTTCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCT
CGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTATCTAC
ACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAGATAGGTGCC
TCACTGATTAAGCATTGGTAATTCGAAATGACCGACCAAGCGACGCCCAACCGGTATCAG
CTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACA
TGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTT
TCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGC
GAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCT
CTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCG
TGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCA
AGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACT
ATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTA
ACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTA
ACTACGGCTACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCT
TCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTT
TTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATTTCAAGAAGATCCTTTGA
TCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCA
TGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTATAGTCCGGAAATACAGGAACGC
ACGCTGGATGGCCCTTCGCTGGGATGGTGAAACCATGAAAAATGGCAGCTTCAGTGGATT
AAGTGGGGGTAATGTGGCCTGTACCCTCTGGTTGCATAGGTATTCATACGGTTAAAATTT
ATCAGGCGCGATTGCGGCAGTTTTTCGGGTGGTTTGTTGCCATTTTTACCTGTCTGCTGC
CGTGATCGCGCTGAACGCGTTTTAGCGGTGCGTACAATTAAGGGATTATGGTAAATCCAC
TTACTGTCTGCCCTCGTAGCCATCGAGATAAACCGCAGTACTCCGGCCACGATGCGTCCG
GCGTAGAGGATCGAGATCT
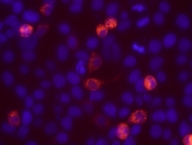
 Characterization of the cloned ORF
Characterization of the cloned ORF N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand
N-terminal HaloTag-fusions expressed in HEK293 visualized by HaloTag TMR ligand Length: 1014 bp
Length: 1014 bp Restriction map A
Restriction map A
 Restriction map B
Restriction map B Genomic Structure
Genomic Structure Length: 338 aa
Length: 338 aa
 Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)
Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)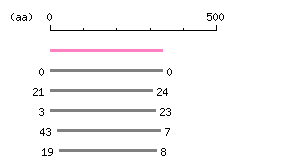
 Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]
Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]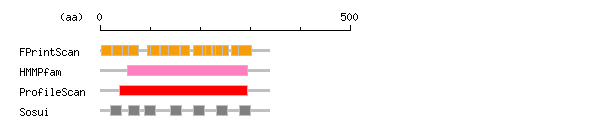
 Result of InterProScan
Result of InterProScan Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)
Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)


 more Linker info
more Linker info