Nucleotide Sequence (with vector) for pF1KB8823
Download
>pF1KB8823 4325 bp
GGATCTCGATCCCGCGAAATTAATACGACTCACTATAGGGGAATTGTGAGCGGATAACAA
TTCCCCACTAGTAATAATTTTCTTTAACTTTAGTAAGGAGCGATCGCCATGTCAGTGAAA
CCCAGCTGGGGGCCTGGCCCCTCGGAGGGGGTCACCGCAGTGCCTACCAGTGACCTTGGA
GAGATCCACAACTGGACCGAGCTGCTTGACCTCTTCAACCACACTTTGTCTGAGTGCCAC
GTGGAGCTCAGCCAGAGCACCAAGCGCGTGGTCCTCTTTGCCCTCTACCTGGCCATGTTT
GTGGTTGGGCTGGTGGAGAACCTCCTGGTGATATGCGTCAACTGGCGCGGCTCAGGCCGG
GCAGGGCTGATGAACCTCTACATCCTCAACATGGCCATCGCGGACCTGGGCATTGTCCTG
TCTCTGCCCGTGTGGATGCTGGAGGTCACGCTGGACTACACCTGGCTCTGGGGCAGCTTC
TCCTGCCGCTTCACTCACTACTTCTACTTTGTCAACATGTATAGCAGCATCTTCTTCCTG
GTGTGCCTCAGTGTCGACCGCTATGTCACCCTCACCAGCGCCTCCCCCTCCTGGCAGCGT
TACCAGCACCGAGTGCGGCGGGCCATGTGTGCAGGCATCTGGGTCCTCTCGGCCATCATC
CCGCTGCCTGAGGTGGTCCACATCCAGCTGGTGGAGGGCCCTGAGCCCATGTGCCTCTTC
ATGGCACCTTTTGAAACGTACAGCACCTGGGCCCTGGCGGTGGCCCTGTCCACCACCATC
CTGGGCTTCCTGCTGCCCTTCCCTCTCATCACAGTCTTCAATGTGCTGACAGCCTGCCGG
CTGCGGCAGCCAGGACAACCCAAGAGCCGGCGCCACTGCCTGCTGCTGTGCGCCTACGTG
GCCGTCTTTGTCATGTGCTGGCTGCCCTATCATGTGACCCTGCTGCTGCTCACACTGCAT
GGGACCCACATCTCCCTCCACTGCCACCTGGTCCACCTGCTCTACTTCTTCTATGATGTC
ATTGACTGCTTCTCCATGCTGCACTGTGTCATCAACCCCATCCTTTACAACTTTCTCAGC
CCACACTTCCGGGGCCGGCTCCTGAATGCTGTAGTCCATTACCTTCCTAAGGACCAGACC
AAGGCGGGCACATGCGCCTCCTCTTCCTCCTGTTCCACCCAGCATTCCATCATCATCACC
AAGGGTGATAGCCAGCCTGCTGCAGCAGCCCCCCACCCTGAGCCAAGCCTGAGCTTTCAG
GCACACCATTTGCTTCCAAATACTTCCCCCATCTCTCCCACTCAGCCTCTTACACCCAGC
GTTTAAACGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCA
AGCTGATCCGGCTGCTAACAAAGCCCGAAAGGAAGCTGAGTTGGCTGCTGCCACCGCTGA
GCAATAACTAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGCTGAA
AGGAGGAACTATATCCGGTTCGCTTGCTGTCCATAAAACCGCCCAGTCTAGCTATCGCCA
TGTAAGCCCACTGCAAGCTACCTGCTTTCTCTTTGCGCTTGCGTTTTCCCTTGTCCAGAT
AGCCCAGTAGCTGACATTCATCCGGGGTCAGCACCGTTTCTGCGGACTGGCTTTCTACGT
GTTCCGCTTCCTTTAGCAGCCCTTGCGCCCTGAGTGCTTGCGGCAGCGTGAGCTTCAAAA
GAATTGCCAGCTGGGGCGCCCTCTGGTAAGGTTGGGAAGCCCTGCAAAGTAAACTGGATG
GCTTTCTTGCCGCCAAGGATCTGATGGCGCAGGGGATCAAGATCTGATCAAGAGACAGGA
TGACGGTCGTTTCGCATGCTTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGG
GTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCC
GTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGT
GCCCTGAATGAACTGCAGGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTT
CCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGC
GAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATC
ATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCAC
CAAGCGAAACATCGCATCGAGCGAGCACGCACTCGGATGGAAGCCGGTCTTGTCGATCAG
GATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAG
GCGCGTATGCCGGATGGTGAGGATCTCGTCGTGACTCATGGCGATGCCTGCTTGCCGAAT
ATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCG
GACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAA
TGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCC
TTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGACTCTGGGGTTCGAAATGACCGACC
AAGCGACGCCCAACCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATC
AGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAA
AAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAA
TCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCC
CCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTC
CGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAG
TTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGA
CCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATC
GCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTAC
AGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGGACAGTATTTGGTATCTG
CGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACA
AACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAA
AGGATTTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAA
CTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTT
ATAGTCCGGAAATACAGGAACGCACGCTGGATGGCCCTTCGCTGGGATGGTGAAACCATG
AAAAATGGCAGCTTCAGTGGATTAAGTGGGGGTAATGTGGCCTGTACCCTCTGGTTGCAT
AGGTATTCATACGGTTAAAATTTATCAGGCGCGATTGCGGCAGTTTTTCGGGTGGTTTGT
TGCCATTTTTACCTGTCTGCTGCCGTGATCGCGCTGAACGCGTTTTAGCGGTGCGTACAA
TTAAGGGATTATGGTAAATCCACTTACTGTCTGCCCTCGTAGCCATCGAGATAAACCGCA
GTACTCCGGCCACGATGCGTCCGGCGTAGAGGATCGAGATCTTTTCAGCCTGATACAGAT
TAAATCAGAACGCAGAAGCGGTCTGATAAAACAGAATTTGCCTGGCGGCAGTAGCGCGGT
GGTCCCACCTGACCCCATGCCGAACTCAGAAGTGAAACGCCGTAGCGCCGATGGTAGTGT
GGGGTCTCCCCATGCGAGAGTAGGGAACTGCCAGGCATCAAATAAAACGAAAGGCTCAGT
CGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCCTGAGTAGGA
CAAATCCGCCGGGAGCGGATTTGAACGTTGCGAAGCAACGGCCCGGAGGGTGGCGGGCAG
GACGCCCGCCATAAACTGCCAGGCATCAAATTAAGCAGAAGGCCATCCTGACGGATGGCC
TTTTTGCGTTTCTACAAACTCTTTTGTTTATTTTTCTAAATACATTCAAATATGTATCCG
CTCAT
 Length: 1212 bp
Length: 1212 bp Restriction map A
Restriction map A
 Restriction map B
Restriction map B Genomic Structure
Genomic Structure Length: 404 aa
Length: 404 aa
 Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)
Result of homology search against CCDS protein database (
FASTA output,
Multiple alignment)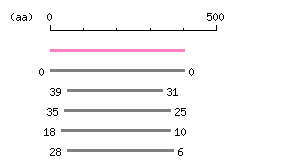
 Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]
Result of motif / domain search (InterProScan and SOSUI)
[ zoom out (x2) ]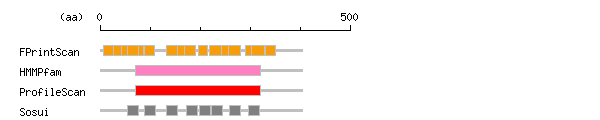
 Result of InterProScan
Result of InterProScan Prediction of transmembrane (TM) segments
Prediction of transmembrane (TM) segments Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)
Result of homology search against disease genes in OMIM database(FASTA output,
Multiple alignment)


 more Linker info
more Linker info