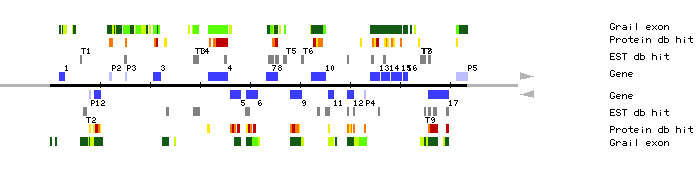
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MSJ1.1 | 1947..2985 | 3 | 0 | giantin | |
| MSJ1.2 | 8883..10148 | 3 | 0 | alternative oxidase AOMI 1 | |
| MSJ1.3 | 20620..22030 | 5 | 0 | hypothetical protein YOR197w | |
| MSJ1.4 | 31693..35499 | 1 | 1 | cosmid T08A11 | |
| MSJ1.5 | 36117..38195 | 4 | 0 | Sequence 1 from Patent WO9534654 | |
| MSJ1.6 | 39282..41426 | 4 | 2 | Sequence 1 from Patent WO9534654 | |
| MSJ1.7 | 43243..44253 | 3 | 1 | GTP cyclohydrolase II (EC 3.5.4.25) / 3,4-dihydroxy-2-butanone 4- phosphate synthase | |
| MSJ1.8 | 44572..45571 | 3 | 2 | GTP cyclohydrolase II (EC 3.5.4.25) | |
| MSJ1.9 | 48034..50223 | 1 | 0 | Sequence of BAC F21M12 from chromosome 1 | |
| MSJ1.10 | 52355..55006 | 6 | 1 | chromosome II BAC T06B20 genomic sequence | |
| MSJ1.11 | 55677..56720 | 1 | 2 | hypothetical 118.6 kD protein c29e6.03c in chromosome I | |
| MSJ1.12 | 59597..60774 | 5 | 5 | immunophilin | |
| MSJ1.13 | 64048..65812 | 6 | 2 | beta-ureidopropionase (EC 3.5.1.6) | |
| MSJ1.14 | 66252..67805 | 4 | 1 | fructose-1,6-bisphosphatase, chloroplast (EC 3.1.3.11) | |
| MSJ1.15 | 68226..70328 | 3 | 0 | hnRNP X protein | |
| MSJ1.16 | 70681..71460 | 3 | 0 | T3 receptor-associating cofactor-1 [human, fetal liver, mRNA, 2930 nt] | |
| MSJ1.17 | 75708..79758 | 5 | 6 | Sequence of BAC F21M12 from chromosome 1 | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MSJ1.P1 | 7925..8008 | 1 | 0 | chloroplast 30 kD ribonucleoprotein precursor | |
| MSJ1.P2 | 62964..63107 | 1 | 0 | K homologue, GrpE homologue, DnaJ homologue and DafA | |
| MSJ1.P3 | 15156..15302 | 1 | 1 | PP1M M110=protein phosphatase 1M 110 kda regulatory subunit [rats, aorta, mRNA, 3300 nt] | |
| MSJ1.P5 | 81200..83594 | 5 | 0 | cosmid B1764 | |
|
|
|||||
|
|
|||||
| identifier | direction | position | Definition of the most similar sequence | ||
|---|---|---|---|---|---|
|
|
|||||
| MSJ1.T1 | 6087..6326 | clone PAP631; 3' end | |||
| MSJ1.T2 | 6724..7275 | clone 106B12T7 | |||
| MSJ1.T3 | 28736..29938 | clone 138J18T7 clone 72B6T7 clone 163H1T7 clone 151G24T7 clone 186J19T7 clone VBVAC07 clone 124K21T7 clone B85XP clone 186E20T7 clone F2B9T7 clone 86G11T7 Lambda-PRL2 cDNA clone 314A7T7 | |||
| MSJ1.T4 | 46732..47333 | clone FAFD25 clone TAT1A4, 5' end clone 141M1T7 clone TAI344 clone TASG047 clone YAP061T clone 193H9T7 | |||
| MSJ1.T5 | 50395..50740 | clone FAFJ56 | |||
| MSJ1.T6 | 74196..74288 | clone 160E8T7 | |||
| MSJ1.T7 | 74408..74594 | clone 163E7T7 clone OAO81; 5' end OS043 NaCl-treated Arabidopsis subtraction library Arabidopsisthaliana cDNA 5' clone 208C14T7 | |||
| MSJ1.T8 | 74866..75149 | CD4-13 cDNA clone E10A5T7 clone OAO81; 3' end | |||
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. III. Sequence Features of the Regions of 1,191,918 bp Covered by Seventeen Physically assigned P1 Clones. (1997) Yasukazu Nakamura, Shusei Sato, Takakazu Kaneko, Hirokazu Kotani, Erika Asamizu, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 401-414
 Kazusa Status
Kazusa Status