|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
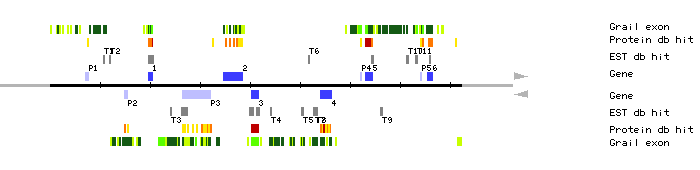
| MRO11.1 | 19649..20551 | 4 | 9 | 60s ribosomal protein L13 | |
| MRO11.2 | 34731..38418 | 12 | 0 | V.radiata for pectinacetylesterase | |
| MRO11.3 | 40205..41768 | 3 | 3 | tubulin beta-8 chain | |
| MRO11.4 | 54003..56269 | 7 | 0 | amino acid permease AAP3 | |
| MRO11.5 | 63133..64544 | 4 | 1 | Rice WSI76 protein induced by water stress | |
| MRO11.6 | 75400..76476 | 6 | 2 | 40s ribosomal protein S11-beta | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MRO11.P1 | 7134..7697 | 2 | 0 | N.tabacum HSR201 protein | |
| MRO11.P2 | 14848..15537 | 2 | 0 | kinesin-like DNA binding protein KID | |
| MRO11.P3 | 26586..32180 | 14 | 3 | cleavage and polyadenylation specificity factor, 100 kd subunit | |
| MRO11.P4 | 62007..62168 | 1 | 0 | zipper protein | |
| MRO11.P5 | 74150..74239 | 1 | 0 | DNA-binding protein | |
|
|
|||||
|
|
|||||
| identifier | direction | position | Definition of the most similar sequence | ||
|---|---|---|---|---|---|
|
|
|||||
| MRO11.T1 | 10628..10943 | clone 205A16T7 clone 127K12T7 | |||
| MRO11.T2 | 11813..12074 | clone GBGF735; 3' end | |||
| MRO11.T3 | 24077..24378 | CD4-15 cDNA clone G12B9T7 | |||
| MRO11.T4 | 44038..44338 | clone 191E18T7 | |||
| MRO11.T5 | 50270..50665 | clone 89N20T7 | |||
| MRO11.T6 | 51740..51975 | clone FAFL75 | |||
| MRO11.T7 | 52601..53004 | clone GBGe302 | |||
| MRO11.T8 | 53244..53376 | clone 127E6T7 | |||
| MRO11.T9 | 66175..66479 | clone YAY223 | |||
| MRO11.T10 | 71264..71650 | clone GBGF846; center | |||
| MRO11.T11 | 73119..73478 | clone 117C22T7 clone 106C12T7 clone 117C23T7 | |||
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. I. Sequence Features of the 1.6 Mb Regions Covered by Physically assigned Twenty P1 Clones. (1997) Shusei Sato, Hirokazu Kotani, Yasukazu Nakamura, Takakazu Kaneko, Erika Asamizu, Masanobu Fukami, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 215-230
 Kazusa Status
Kazusa Status