|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
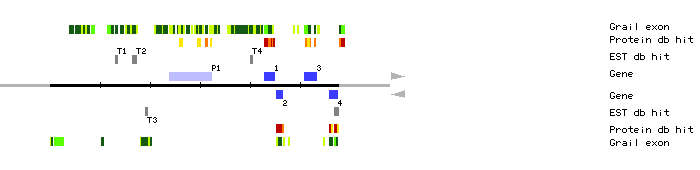
| MOP10.1 | 42946..44957 | 8 | 0 | temperature-sensitive omega-3 fatty acid desaturase, chloroplast precursor (EC 1.14.99.-) | |
| MOP10.2 | 45352..46566 | 4 | 0 | Arabidopsis thaliana phosphoribosylanthranilate isomerase | |
| MOP10.3 | 50881..53214 | 3 | 0 | Picea glauca ethylene-forming enzyme (EFE) | |
| MOP10.4 | 55949..57557 | 4 | 2 | Alfalfa nucleic acid binding protein | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MOP10.P1 | 23933..32358 | 21 | 0 | protein tsg24 | |
|
|
|||||
|
|
|||||
| identifier | direction | position | Definition of the most similar sequence | ||
|---|---|---|---|---|---|
|
|
|||||
| MOP10.T1 | 13007..13499 | clone 180C3T7 | |||
| MOP10.T2 | 16561..17305 | clone 143H16T7 clone 143G16T7 | |||
| MOP10.T3 | 19188..19457 | clone 99P2T7 | |||
| MOP10.T4 | 40070..40579 | clone 166J8T7 | |||
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. I. Sequence Features of the 1.6 Mb Regions Covered by Physically assigned Twenty P1 Clones. (1997) Shusei Sato, Hirokazu Kotani, Yasukazu Nakamura, Takakazu Kaneko, Erika Asamizu, Masanobu Fukami, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 215-230
 Kazusa Status
Kazusa Status