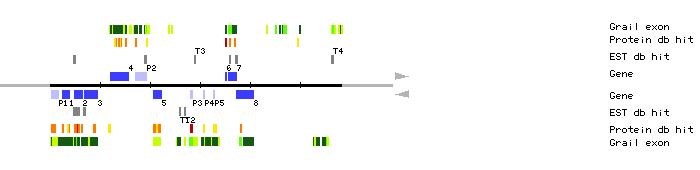
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MLE2.1 | 2404..3867 | 5 | 0 | auxin-independent growth promoter | |
| MLE2.2 | 4931..6471 | 5 | 7 | adenylate kinase b (EC 2.7.4.3) | |
| MLE2.3 | 6830..9422 | 6 | 1 | receptor protein kinase | |
| MLE2.4 | 12169..15627 | 11 | 0 | hypothetical 69.1 kD protein | |
| MLE2.5 | 20644..22347 | 1 | 0 | cytochrome p450 LXXXVI (EC 1.14.-.-) | |
| MLE2.6 | 35114..35320 | 1 | 0 | AT.I.24-1, AT.I.24-2, AT.I.24-3, AT.I.24-4, AT.I.24-5, AT.I.24-6, AT.I.24-9 and AT.I.24-14 genesAT.I.24-7, ascorbate peroxidase and AT.I.24-13 g | |
| MLE2.7 | 35708..37263 | 2 | 3 | ferripyochelin binding protein homolog | |
| MLE2.8 | 37241..40777 | 9 | 2 | hypothetical protein | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MLE2.P1 | 296..1680 | 2 | 0 | luciferase GR | |
| MLE2.P2 | 17070..19392 | 2 | 1 | DNA-binding protein Gt-2 | |
| MLE2.P3 | 28139..28588 | 1 | 0 | F7G19 | |
| MLE2.P4 | 30723..30809 | 1 | 0 | 10 kD chaperonin | |
| MLE2.P5 | 32732..32836 | 1 | 0 | hypothetical protein | |
|
|
|||||
|
|
|||||
| identifier | direction | position | Definition of the most similar sequence | ||
|---|---|---|---|---|---|
|
|
|||||
| MLE2.T1 | 25856..26004 | clone 169N24T7 | |||
| MLE2.T2 | 26930..27126 | clone 94M10T7 | |||
| MLE2.T3 | 28961..29051 | clone GBGa344, 5' end | |||
| MLE2.T4 | 56393..56684 | clone 41D8T7 | |||
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. III. Sequence Features of the Regions of 1,191,918 bp Covered by Seventeen Physically assigned P1 Clones. (1997) Yasukazu Nakamura, Shusei Sato, Takakazu Kaneko, Hirokazu Kotani, Erika Asamizu, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 401-414
 Kazusa Status
Kazusa Status