|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
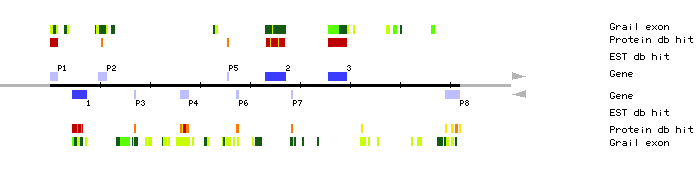
| MIK22.1 | 4549..7353 | 8 | 0 | A.thaliana glucose-6-phosphate dehydrogenase | |
| MIK22.2 | 43150..47175 | 1 | 0 | retrovirus-related polyprotein | |
| MIK22.3 | 55716..59283 | 3 | 0 | phytochrome c | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MIK22.P1 | 8..1417 | 1 | 0 | Zea mays copia-like retrotransposon Hopscotch polyprotein | |
| MIK22.P2 | 9699..11267 | 2 | 0 | mudrA protein | |
| MIK22.P3 | 16850..17140 | 1 | 0 | S-receptor kinase (EC 2.7.1.-) 8 precursor | |
| MIK22.P4 | 26123..27730 | 4 | 0 | A.thaliana 81kb genomic sequence | |
| MIK22.P5 | 35466..35561 | 1 | 0 | phytochrome c | |
| MIK22.P6 | 37332..37691 | 1 | 0 | reverse transcriptase | |
| MIK22.P7 | 48284..48484 | 1 | 0 | glutaminase, kidney isoform precursor (EC 3.5.1.2) | |
| MIK22.P8 | 79164..81955 | 8 | 0 | autoantigen pm-scl | |
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. I. Sequence Features of the 1.6 Mb Regions Covered by Physically assigned Twenty P1 Clones. (1997) Shusei Sato, Hirokazu Kotani, Yasukazu Nakamura, Takakazu Kaneko, Erika Asamizu, Masanobu Fukami, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 215-230
 Kazusa Status
Kazusa Status