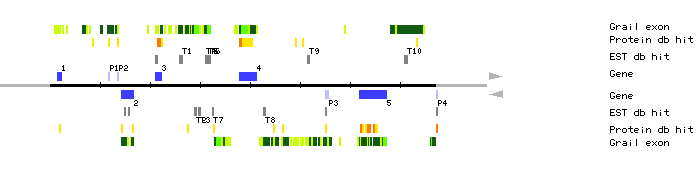
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MDJ22.1 | 1496..2313 | 2 | 0 | cell surface-binding protein | |
| MDJ22.2 | 14212..16701 | 7 | 2 | Synechocystis sp. PCC6803 slr1124 (phosphoglycerate mutase) | |
| MDJ22.3 | 21123..22397 | 1 | 1 | prephenate dehydratase (EC 4.2.1.51) | |
| MDJ22.4 | 37847..41363 | 5 | 0 | TMV resistance protein N | |
| MDJ22.5 | 61855..67349 | 20 | 0 | dna repair protein rad8 | |
|
|
|||||
|
|
|||||
| identifier | direction | position | No. of exon |
No. of EST |
Definition of the most similar sequence |
|---|---|---|---|---|---|
|
|
|||||
| MDJ22.P1 | 11677..11887 | 1 | 0 | Caenorhabditis elegans cosmid K04A8 | |
| MDJ22.P2 | 13458..13619 | 1 | 0 | ||
| MDJ22.P3 | 55112..55694 | 2 | 1 | Acetobacter xylinum cellulose synthase | |
| MDJ22.P4 | 77255..77363 | 1 | 1 | alpha-adaptin | |
|
|
|||||
|
|
|||||
| identifier | direction | position | Definition of the most similar sequence | ||
|---|---|---|---|---|---|
|
|
|||||
| MDJ22.T1 | 25909..26443 | clone TAP0356; 5' end CD4-16 cDNA clone H1C10T7 | |||
| MDJ22.T2 | 28969..29332 | clone TAP0356; 3' end | |||
| MDJ22.T3 | 29715..30189 | CD4-16 cDNA clone H9H3T7 | |||
| MDJ22.T4 | 31030..31244 | clone TAT6B01; 5' end; Similar toATTS0396 clone 133F15T7 | |||
| MDJ22.T5 | 31535..31712 | clone 117P21T7 | |||
| MDJ22.T6 | 31819..32105 | clone FAI214; 5' end clone TAT4F5, 5' end | |||
| MDJ22.T7 | 32469..32732 | clone TAT6B01; 3' end | |||
| MDJ22.T8 | 42666..43078 | Lambda-PRL2 cDNA clone 200M20T7 clone 183D14T7 | |||
| MDJ22.T9 | 51596..51896 | clone FAFL51; 3' end | |||
| MDJ22.T10 | 70907..71588 | CD4-16 cDNA clone H10F9T7 | |||
|
|
|||||
Structural Analysis of Arabidopsis thaliana Chromosome 5. II. Sequence Features of the Regions of 1,044,062 bp Covered by Thirteen Physically assigned P1 Clones. (1997) Hirokazu Kotani, Yasukazu Nakamura, Shusei Sato, Takakazu Kaneko, Erika Asamizu, Nobuyuki Miyajima, and Satoshi Tabata DNA Research, 4, 291-300
 Kazusa Status
Kazusa Status