Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002328A_C01 KMC002328A_c01
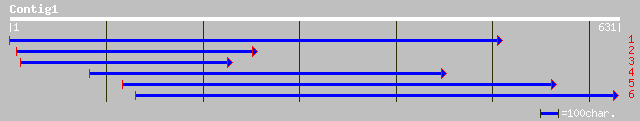
(623 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196901.1| putative protein; protein id: At5g13970.1, supp... 55 1e-06
gb|EAA21238.1| hypothetical protein [Plasmodium yoelii yoelii] 39 0.055
gb|EAA28084.1| hypothetical protein [Neurospora crassa] 32 0.44
ref|XP_230343.1| similar to ribosomal protein L7a, cytosolic [va... 36 0.47
ref|XP_243736.1| hypothetical protein XP_243736 [Rattus norvegicus] 35 0.80
>ref|NP_196901.1| putative protein; protein id: At5g13970.1, supported by cDNA:
gi_19310485 [Arabidopsis thaliana]
gi|10177663|dbj|BAB11125.1|
emb|CAB86638.1~gene_id:MAC12.6~similar to unknown
protein [Arabidopsis thaliana]
gi|19310486|gb|AAL84977.1| AT5g13970/MAC12_6
[Arabidopsis thaliana] gi|28416455|gb|AAO42758.1|
At5g13970/MAC12_6 [Arabidopsis thaliana]
Length = 404
Score = 54.7 bits (130), Expect = 1e-06
Identities = 35/122 (28%), Positives = 61/122 (49%), Gaps = 1/122 (0%)
Frame = -2
Query: 622 ASDMDDKANKEAYMGFLAQMKGTGSQADDALDDLPTSSPFISKKKSSDVTMVENEMVSRQ 443
+ ++D+++N++AYM FL ++ D L +LP S F+ K+K + VEN
Sbjct: 296 SGEVDEESNRKAYMDFLNMIRSKDESLVDPLMELPRSVAFVPKRKPMAESKVEN------ 349
Query: 442 KLDVGKESMNKKAFPVSIAAAGDN-ENSDVCAMEEDEPEVMEDAKKSSQRLNNRKYRKKP 266
+ K+ ++ +A A D E+ + AMEEDEPE + K R + ++ P
Sbjct: 350 ---IDKDCEGRR-----VAIAVDTIEDCTISAMEEDEPETAQHVTKRPGRQYRARAKEDP 401
Query: 265 QE 260
+E
Sbjct: 402 EE 403
>gb|EAA21238.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 957
Score = 38.9 bits (89), Expect = 0.055
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 9/103 (8%)
Frame = -2
Query: 541 DDALDDL-PTSSPFISKKKSSDVTMVENEMVSRQKL---DVGKESMNKKAFPVSIAAAGD 374
DD +++ P ++P + KKK+ + V+ QK+ D K ++N+ P+ D
Sbjct: 21 DDPKNEVNPNATPNVKKKKNKNTNQVDKSNKKSQKIIVEDTNKTNINESNNPIKTIEIND 80
Query: 373 NE---NSDVCAMEE--DEPEVMEDAKKSSQRLNNRKYRKKPQE 260
NE +D+ A D P ++ +R NN+K + K E
Sbjct: 81 NEETKTNDLSAQNSKLDTPNKNDNKTNLKKRKNNKKEKLKLNE 123
>gb|EAA28084.1| hypothetical protein [Neurospora crassa]
Length = 1027
Score = 32.0 bits (71), Expect(2) = 0.44
Identities = 17/36 (47%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Frame = +3
Query: 165 KGTKKYTSQKSEIGKLCKSRNSFCGY-TTCSSNSWG 269
+ T TS +S L +SR+S G TTCSS SWG
Sbjct: 41 QSTNSTTSTRSGPASLARSRSSTAGASTTCSSRSWG 76
Score = 22.7 bits (47), Expect(2) = 0.44
Identities = 7/12 (58%), Positives = 8/12 (66%)
Frame = +1
Query: 337 HPPPLHKHHCFH 372
HP P H+HH H
Sbjct: 109 HPHPHHRHHHHH 120
>ref|XP_230343.1| similar to ribosomal protein L7a, cytosolic [validated] - rat
[Rattus norvegicus]
Length = 415
Score = 35.8 bits (81), Expect = 0.47
Identities = 22/86 (25%), Positives = 40/86 (45%), Gaps = 1/86 (1%)
Frame = -2
Query: 505 FISKKKS-SDVTMVENEMVSRQKLDVGKESMNKKAFPVSIAAAGDNENSDVCAMEEDEPE 329
FI ++++ S V +E + + L + + G E +D EE+ E
Sbjct: 49 FIQEQRTESKVRAIEKGQAALEMLQEERSEKQGSSKETDSEKTGTEEETDETDSEEELQE 108
Query: 328 VMEDAKKSSQRLNNRKYRKKPQELEE 251
+++ +K S R +K RKKP+ +EE
Sbjct: 109 LIDQMRKQSLRNRQKKNRKKPKMVEE 134
>ref|XP_243736.1| hypothetical protein XP_243736 [Rattus norvegicus]
Length = 175
Score = 35.0 bits (79), Expect = 0.80
Identities = 26/103 (25%), Positives = 48/103 (46%)
Frame = -2
Query: 559 GTGSQADDALDDLPTSSPFISKKKSSDVTMVENEMVSRQKLDVGKESMNKKAFPVSIAAA 380
GT S+ + P+ ++K + + M++ E +Q KE+ ++K
Sbjct: 36 GTISEICTTRAEFPSPQQLYAEKGQTALEMLQEERSEKQ--GSAKETDSEKT-------- 85
Query: 379 GDNENSDVCAMEEDEPEVMEDAKKSSQRLNNRKYRKKPQELEE 251
E +D EE+ E+++ +K S R +K RKKP+ +EE
Sbjct: 86 DTEEETDETDSEEELQELIDQMQKQSLRNRQKKKRKKPKMVEE 128
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,670,333
Number of Sequences: 1393205
Number of extensions: 11667036
Number of successful extensions: 35201
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 33021
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35044
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)