Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002184A_C01 KMC002184A_c01
(979 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
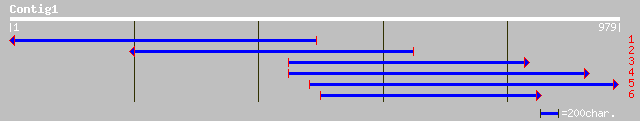
pir||S51910 cryptogene protein G4 - Leishmania tarentolae (strai... 40 0.055
gb|AAM44364.1| POTASSIUM CHANNEL REGULATORY FACTOR [Dictyosteliu... 40 0.055
gb|AAC38978.1| cytochrome c oxidase subunit 1 [Bodo saltans] 35 1.4
ref|NP_694904.1| NADH dehydrogenase subunit 4 [Brugia malayi] gi... 35 2.3
ref|NP_703268.1| hypothetical protein [Plasmodium falciparum 3D7... 34 3.0
>pir||S51910 cryptogene protein G4 - Leishmania tarentolae (strain LEM125)
Length = 169
Score = 40.0 bits (92), Expect = 0.055
Identities = 47/184 (25%), Positives = 74/184 (39%), Gaps = 7/184 (3%)
Frame = -1
Query: 667 EYCTIFYFLGFLNMVILD*WATNCPINSPLICFCCISFFCFTALLPN*LRINYFFLYLRF 488
+ C + YF+ ++ V L + C + L+CF CI FF FT + I +FFL++ +
Sbjct: 32 DLCVMVYFI-YIAFVCL--FVLVCYVVILLLCFDCIVFFLFTVI------IIFFFLFIFW 82
Query: 487 IMKNHGRRIEKVSKILILL-------LSSFFYKPN*LPINSFLLHFLFLFHCFKTCVWFM 329
+ I +LL L FFY + + +F FLF F F+
Sbjct: 83 VF------------IFLLLFWFVFGFLFFFFY------VCVLIFYFEFLFMLFFVFCGFL 124
Query: 328 LF*VF*HLI*AGGTL**LKFGC*K**FYAVFIVM**AVVFYAVFICSFQMFLL*YCNRCL 149
LF +F F+ F V+ V + +FI F+ F + +C L
Sbjct: 125 LFVMFIL-------------------FFISFFVLIVLVFCWLLFIFVFRFFCMTFCILIL 165
Query: 148 ISCF 137
I F
Sbjct: 166 IGLF 169
>gb|AAM44364.1| POTASSIUM CHANNEL REGULATORY FACTOR [Dictyostelium discoideum]
gi|28828386|gb|AAO51022.1| similar to Homo sapiens
(Human). HPRP18 (Pre-mRNA splicing factor similar to S.
CEREVISIAE PRP18) [Dictyostelium discoideum]
Length = 389
Score = 40.0 bits (92), Expect = 0.055
Identities = 24/72 (33%), Positives = 40/72 (55%)
Frame = +2
Query: 170 EQKHLK*TNKNCIKNNSLSHNNKNCIKSSFLTTKF*LLKGATSSNQMSKDLKQHKPNTRL 349
++ LK +N N NN+ S++N C KS TK +G ++ K+LKQ K R+
Sbjct: 16 KEAQLKNSNNNNNNNNNNSNSNNGCDKSEEPATKKYKTRG-----EIDKELKQKKEEERI 70
Query: 350 KTVKQKKEMQQK 385
K K+K++ ++K
Sbjct: 71 KKEKEKEKEKEK 82
>gb|AAC38978.1| cytochrome c oxidase subunit 1 [Bodo saltans]
Length = 548
Score = 35.4 bits (80), Expect = 1.4
Identities = 23/82 (28%), Positives = 43/82 (52%), Gaps = 7/82 (8%)
Frame = -3
Query: 386 VFVAFPFSVSLF*DVCLVYVVLSLLT----FDLSWWHPLIVKIWLLKM---MILCSFYCY 228
+F A FSV +F D C+ +V + + + + W P ++ ++LL + I+ +
Sbjct: 468 LFAACLFSVLMFWDYCIFFVSIFIYCLFCFYSFNSWLPFVMALYLLLVDFAHIVLDYLFI 527
Query: 227 VISCCFLCSFYLFISNVSALVL 162
+IS CF+ +YLF + L+L
Sbjct: 528 IISFCFI--YYLFFFSNFVLLL 547
>ref|NP_694904.1| NADH dehydrogenase subunit 4 [Brugia malayi]
gi|23307677|gb|AAN17805.1| NADH dehydrogenase subunit 4
[Brugia malayi]
Length = 409
Score = 34.7 bits (78), Expect = 2.3
Identities = 31/115 (26%), Positives = 48/115 (40%), Gaps = 5/115 (4%)
Frame = -1
Query: 652 FYFLGFLNMVILD*WATNCPINSPLICFCCISFFCFTALLPN*LRINYFFLYLR-----F 488
F FL F+++ IL + + + C I FF I Y F L F
Sbjct: 47 FVFLSFMSVFILGFICVSEVLGGLVFYSCLIVFFSVCFFFSGSFLILYIFYELTMLPVLF 106
Query: 487 IMKNHGRRIEKVSKILILLLSSFFYKPN*LPINSFLLHFLFLFHCFKTCVWFMLF 323
+ +GR++EK+S L+ + F+ +P F H +F F F +FM F
Sbjct: 107 CLLGYGRQVEKISACYYLVFYTLFFG---MPFLFFFSH-VFNFFNFVYYDFFMSF 157
>ref|NP_703268.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23510640|emb|CAD49025.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 299
Score = 34.3 bits (77), Expect = 3.0
Identities = 21/71 (29%), Positives = 32/71 (44%)
Frame = +2
Query: 173 QKHLK*TNKNCIKNNSLSHNNKNCIKSSFLTTKF*LLKGATSSNQMSKDLKQHKPNTRLK 352
+K + NK +N + + NKN K K + N+ K LK+ K +
Sbjct: 68 RKRKRRRNKRTRRNRTQARKNKNKRKKKKKNEKLAKRRRRRERNRKKKQLKKEKRRRKKN 127
Query: 353 TVKQKKEMQQK 385
KQKKEM++K
Sbjct: 128 EKKQKKEMKRK 138
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,749,020
Number of Sequences: 1393205
Number of extensions: 16467102
Number of successful extensions: 44739
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 40165
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44458
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 56019656464
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)