Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000477A_C01 KMC000477A_c01
(592 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
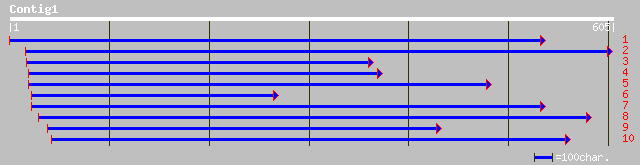
pir||T50775 probable translation initiation factor [imported] - ... 141 7e-33
sp|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation factor 3 ... 140 1e-32
pir||T50773 translation initiation factor homolog p105 [imported... 72 5e-12
ref|NP_191174.1| PROBABLE EUKARYOTIC TRANSLATION INITIATION FACT... 72 5e-12
dbj|BAC56812.1| putative Eukaryotic translation initiation facto... 69 4e-11
>pir||T50775 probable translation initiation factor [imported] - barrel medic
gi|5106928|gb|AAD39892.1|AF106931_1 putative translation
initiation protein [Medicago truncatula]
Length = 383
Score = 141 bits (355), Expect = 7e-33
Identities = 81/112 (72%), Positives = 88/112 (78%), Gaps = 4/112 (3%)
Frame = -2
Query: 591 ERAAEARIGGGGLDLPPRRRDGQDYAAAAAGGGGG--AGGRWQDLSLSQPRQGSGRAGYG 418
ERA+EAR+GGGGLDLPPRRRDGQDYAAAAAGGGGG +GGRWQDLS SQ RQGSGRAGY
Sbjct: 267 ERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY- 325
Query: 417 SGGGRPFAFNQA--AGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQGGSALQG 268
GGR +FNQA +GGYSR R G GGGGYQ+ S R QGGSAL+G
Sbjct: 326 --GGRALSFNQAGGSGGYSRGR----GMGGGGYQN----SSRT-QGGSALRG 366
>sp|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110)
(eIF3c) gi|11282366|pir||T50774 probable translation
initiation factor [imported] - barrel medic
gi|5106926|gb|AAD39891.1|AF106930_1 putative translation
initiation protein [Medicago truncatula]
Length = 935
Score = 140 bits (353), Expect = 1e-32
Identities = 80/112 (71%), Positives = 87/112 (77%), Gaps = 4/112 (3%)
Frame = -2
Query: 591 ERAAEARIGGGGLDLPPRRRDGQDYAAAAAGGGGG--AGGRWQDLSLSQPRQGSGRAGYG 418
ERA EAR+GGGGLDLPPRRRDGQDYAAAAAGGG G +GGRWQDLS SQ RQGSGR GY
Sbjct: 818 ERATEARLGGGGLDLPPRRRDGQDYAAAAAGGGSGTSSGGRWQDLSYSQTRQGSGRTGY- 876
Query: 417 SGGGRPFAFNQA--AGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQGGSALQG 268
GGGR +F+QA +GGYSR R G GGGGYQ+ SGR QGGSAL+G
Sbjct: 877 -GGGRALSFSQAGGSGGYSRGR----GTGGGGYQN----SGRT-QGGSALRG 918
>pir||T50773 translation initiation factor homolog p105 [imported] - Arabidopsis
thaliana gi|2789660|gb|AAC83464.1| eIF3c [Arabidopsis
thaliana]
Length = 900
Score = 72.0 bits (175), Expect = 5e-12
Identities = 50/103 (48%), Positives = 57/103 (54%), Gaps = 2/103 (1%)
Frame = -2
Query: 591 ERAAEARIGGGGLDLPPRRRD-GQDYAAAAAGGGGGAGGRWQD-LSLSQPRQGSGRAGYG 418
ERA E+R GGGGLDL RRRD QDYA AA+GG GG WQD + Q RQG+ R+GY
Sbjct: 798 ERAMESRTGGGGLDLSSRRRDNNQDYAGAASGG----GGYWQDKANYGQGRQGN-RSGY- 851
Query: 417 SGGGRPFAFNQAAGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQ 289
GGGR N G +R GGGY + G R Q
Sbjct: 852 -GGGRSSGQNGQWSGQNR---------GGGYAGRVGSGNRGMQ 884
>ref|NP_191174.1| PROBABLE EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT 8;
protein id: At3g56150.1, supported by cDNA: gi_17381099,
supported by cDNA: gi_2789659 [Arabidopsis thaliana]
gi|23503063|sp|O49160|IF38_ARATH Eukaryotic translation
initiation factor 3 subunit 8 (eIF3 p110) (eIF3c) (p105)
gi|11282364|pir||T47732 probable translation initiation
factor eIF-3 chain 8 F18O21.110 [imported] - Arabidopsis
thaliana gi|7572913|emb|CAB87414.1| PROBABLE EUKARYOTIC
TRANSLATION INITIATION FACTOR 3 SUBUNIT 8 [Arabidopsis
thaliana] gi|17381100|gb|AAL36362.1| putative eukaryotic
translation initiation factor 3 subunit 8 [Arabidopsis
thaliana] gi|25054977|gb|AAN71960.1| putative eukaryotic
translation initiation factor 3 subunit 8 [Arabidopsis
thaliana]
Length = 900
Score = 72.0 bits (175), Expect = 5e-12
Identities = 50/103 (48%), Positives = 57/103 (54%), Gaps = 2/103 (1%)
Frame = -2
Query: 591 ERAAEARIGGGGLDLPPRRRD-GQDYAAAAAGGGGGAGGRWQD-LSLSQPRQGSGRAGYG 418
ERA E+R GGGGLDL RRRD QDYA AA+GG GG WQD + Q RQG+ R+GY
Sbjct: 798 ERAMESRTGGGGLDLSSRRRDNNQDYAGAASGG----GGYWQDKANYGQGRQGN-RSGY- 851
Query: 417 SGGGRPFAFNQAAGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQ 289
GGGR N G +R GGGY + G R Q
Sbjct: 852 -GGGRSSGQNGQWSGQNR---------GGGYAGRVGSGNRGMQ 884
>dbj|BAC56812.1| putative Eukaryotic translation initiation factor 3 subunit 8 (eIF3
p110) [Oryza sativa (japonica cultivar-group)]
Length = 936
Score = 68.9 bits (167), Expect = 4e-11
Identities = 50/113 (44%), Positives = 61/113 (53%), Gaps = 5/113 (4%)
Frame = -2
Query: 591 ERAAEARIGGGGLDLPPRRR--DGQDYAAAAAGGGGGAGGRWQDLSLS-QPRQGSGRAGY 421
ERA EA+ GG PPRRR DGQD + G+WQ+ +S Q RQG GR+GY
Sbjct: 821 ERAYEAKTGGTLEGAPPRRRGGDGQDSSNL---------GKWQENFVSSQGRQGGGRSGY 871
Query: 420 GSGGGRPFAFNQAAGGYSRDRTGRG--GGGGGGYQSQYGGSGRAHQGGSALQG 268
G P + GGY RDR +G GG GGG + Q GG R +Q GS +G
Sbjct: 872 SGRVGGP---GRGGGGYQRDRGSQGSRGGYGGGSRFQDGGRSR-NQSGSMARG 920
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 585,688,785
Number of Sequences: 1393205
Number of extensions: 18096777
Number of successful extensions: 313362
Number of sequences better than 10.0: 5690
Number of HSP's better than 10.0 without gapping: 83164
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 175300
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)