Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC020557A_C01 KMC020557A_c01
(588 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
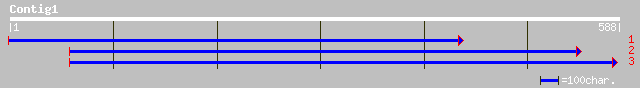
gb|AAD32302.1| ribonucleotide reductase small subunit [Glycine max] 244 4e-64
ref|NP_189000.1| ribonucleoside-diphosphate reductase small chai... 207 6e-53
pir||S68538 ribonucleoside-diphosphate reductase (EC 1.17.4.1) s... 201 4e-51
sp|P49730|RIR2_TOBAC Ribonucleoside-diphosphate reductase small ... 198 4e-50
ref|NP_189342.1| ribonucleotide reductase small subunit, putativ... 192 2e-48
>gb|AAD32302.1| ribonucleotide reductase small subunit [Glycine max]
Length = 339
Score = 244 bits (624), Expect = 4e-64
Identities = 117/130 (90%), Positives = 126/130 (96%)
Frame = -3
Query: 586 RDEGLHCDFACLLYSLLQRPLDSERVHHIVHEAVEIETEFVCEALPCALIGMNSTLMSQY 407
RDEGLHCDFACLLYSLL++PL S++VH +VHEAVEIETEFVC+ALPCALIGMNS LMSQY
Sbjct: 210 RDEGLHCDFACLLYSLLRKPLISDQVHKLVHEAVEIETEFVCDALPCALIGMNSVLMSQY 269
Query: 406 IKFVADRLLVSLGYQKKYNVGNPFDWMEFISLQGKANFFERRVGEYQKASVMSSLQDAGK 227
IKFVADRLLV+LGYQ+KYNV NPFDWMEFISLQGKANFFERRVG+YQKASVMSSLQDAGK
Sbjct: 270 IKFVADRLLVALGYQRKYNVENPFDWMEFISLQGKANFFERRVGDYQKASVMSSLQDAGK 329
Query: 226 NFVFKLDEDF 197
NFVFKLDEDF
Sbjct: 330 NFVFKLDEDF 339
>ref|NP_189000.1| ribonucleoside-diphosphate reductase small chain; protein id:
At3g23580.1, supported by cDNA: gi_13937231 [Arabidopsis
thaliana] gi|27735240|sp|P50651|RIR2_ARATH
Ribonucleoside-diphosphate reductase small chain
(Ribonucleotide reductase) (R2 subunit)
gi|9294514|dbj|BAB02776.1| ribonucleotide reductase
[Arabidopsis thaliana]
gi|13937232|gb|AAK50108.1|AF372971_1 AT3g23580/MDB19_7
[Arabidopsis thaliana] gi|23505945|gb|AAN28832.1|
At3g23580/MDB19_7 [Arabidopsis thaliana]
Length = 341
Score = 207 bits (528), Expect = 6e-53
Identities = 99/130 (76%), Positives = 113/130 (86%)
Frame = -3
Query: 586 RDEGLHCDFACLLYSLLQRPLDSERVHHIVHEAVEIETEFVCEALPCALIGMNSTLMSQY 407
RDEGLHCDFACLLYSLLQ+ L E+V+ IVHEAVEIETEFVC+ALPC LIGMNS LMSQY
Sbjct: 212 RDEGLHCDFACLLYSLLQKQLPLEKVYQIVHEAVEIETEFVCKALPCDLIGMNSNLMSQY 271
Query: 406 IKFVADRLLVSLGYQKKYNVGNPFDWMEFISLQGKANFFERRVGEYQKASVMSSLQDAGK 227
I+FVADRLLV+LG ++ Y NPFDWMEFISLQGK NFFE+RVGEYQKASVMS+LQ+ +
Sbjct: 272 IQFVADRLLVTLGCERTYKAENPFDWMEFISLQGKTNFFEKRVGEYQKASVMSNLQNGNQ 331
Query: 226 NFVFKLDEDF 197
N+ F +EDF
Sbjct: 332 NYEFTTEEDF 341
>pir||S68538 ribonucleoside-diphosphate reductase (EC 1.17.4.1) small chain -
Arabidopsis thaliana gi|840719|emb|CAA54549.1|
ribonucleotide reductase R2 [Arabidopsis thaliana]
Length = 340
Score = 201 bits (512), Expect = 4e-51
Identities = 97/130 (74%), Positives = 111/130 (84%)
Frame = -3
Query: 586 RDEGLHCDFACLLYSLLQRPLDSERVHHIVHEAVEIETEFVCEALPCALIGMNSTLMSQY 407
RDEGLHCDFACLL SLLQ + E+V+ IVHEAVEIETEFVC+ALPC LIGMNS LMSQY
Sbjct: 211 RDEGLHCDFACLLISLLQLHVPLEKVYQIVHEAVEIETEFVCKALPCDLIGMNSNLMSQY 270
Query: 406 IKFVADRLLVSLGYQKKYNVGNPFDWMEFISLQGKANFFERRVGEYQKASVMSSLQDAGK 227
I+FVADRLLV+LG ++ Y NPFDWMEFISLQGK NFFE+RVGEYQKASVMS+LQ+ +
Sbjct: 271 IQFVADRLLVTLGCERTYKAENPFDWMEFISLQGKTNFFEKRVGEYQKASVMSNLQNGNQ 330
Query: 226 NFVFKLDEDF 197
N+ F +EDF
Sbjct: 331 NYEFTTEEDF 340
>sp|P49730|RIR2_TOBAC Ribonucleoside-diphosphate reductase small chain (Ribonucleotide
reductase) (R2 subunit) gi|7433357|pir||T03688
ribonucleoside-diphosphate reductase (EC 1.17.4.1) chain
R2 - common tobacco gi|1044912|emb|CAA63194.1|
ribonucleotide reductase R2 [Nicotiana tabacum]
Length = 329
Score = 198 bits (504), Expect = 4e-50
Identities = 98/130 (75%), Positives = 106/130 (81%)
Frame = -3
Query: 586 RDEGLHCDFACLLYSLLQRPLDSERVHHIVHEAVEIETEFVCEALPCALIGMNSTLMSQY 407
RDEGLHCDFACLLYSLL+ L ERV IV +AVEIE EFVC+ALPCAL+GMN LMS+Y
Sbjct: 200 RDEGLHCDFACLLYSLLRTKLTEERVKGIVADAVEIEREFVCDALPCALVGMNGDLMSKY 259
Query: 406 IKFVADRLLVSLGYQKKYNVGNPFDWMEFISLQGKANFFERRVGEYQKASVMSSLQDAGK 227
I+FVADRLL +LGY K YN NPFDWME ISLQGK NFFE+RVGEYQKASVMSSL G
Sbjct: 260 IEFVADRLLDALGYDKLYNAQNPFDWMELISLQGKTNFFEKRVGEYQKASVMSSLNGNGA 319
Query: 226 NFVFKLDEDF 197
FKLDEDF
Sbjct: 320 THEFKLDEDF 329
>ref|NP_189342.1| ribonucleotide reductase small subunit, putative; protein id:
At3g27060.1, supported by cDNA: gi_17380725 [Arabidopsis
thaliana] gi|9279629|dbj|BAB01087.1| ribonucleotide
reductase [Arabidopsis thaliana]
gi|17380726|gb|AAL36193.1| putative ribonucleotide
reductase small subunit [Arabidopsis thaliana]
gi|21436297|gb|AAM51287.1| putative ribonucleotide
reductase small subunit [Arabidopsis thaliana]
Length = 332
Score = 192 bits (489), Expect = 2e-48
Identities = 97/132 (73%), Positives = 107/132 (80%), Gaps = 2/132 (1%)
Frame = -3
Query: 586 RDEGLHCDFACLLYSLLQRPLDSERVHHIVHEAVEIETEFVCEALPCALIGMNSTLMSQY 407
RDEGLHCDFACLLY+LL+ L ERV IV +AVEIE EFVC+ALPCAL+GMN LMSQY
Sbjct: 201 RDEGLHCDFACLLYTLLKTKLSEERVKSIVCDAVEIEREFVCDALPCALVGMNRDLMSQY 260
Query: 406 IKFVADRLLVSLGYQKKYNVGNPFDWMEFISLQGKANFFERRVGEYQKASVMSSLQDAG- 230
I+FVADRLL +LGY K Y V NPFDWME ISLQGK NFFE+RVG+YQKASVMSS+ G
Sbjct: 261 IEFVADRLLGALGYGKVYGVTNPFDWMELISLQGKTNFFEKRVGDYQKASVMSSVNGNGA 320
Query: 229 -KNFVFKLDEDF 197
N VF LDEDF
Sbjct: 321 FDNHVFSLDEDF 332
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,847,053
Number of Sequences: 1393205
Number of extensions: 10867824
Number of successful extensions: 26204
Number of sequences better than 10.0: 197
Number of HSP's better than 10.0 without gapping: 25413
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26129
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)