Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC020239A_C01 KMC020239A_c01
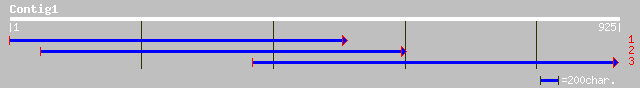
(925 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181947.1| hypothetical protein; protein id: At2g44190.1 [... 177 2e-43
ref|NP_191559.1| putative protein; protein id: At3g60000.1 [Arab... 164 2e-39
dbj|BAC42908.1| unknown protein [Arabidopsis thaliana] gi|289732... 133 3e-30
ref|NP_564558.1| expressed protein; protein id: At1g49890.1, sup... 125 1e-27
pir||T52384 hypothetical protein MMB12.3 [imported] - Arabidopsi... 120 4e-26
>ref|NP_181947.1| hypothetical protein; protein id: At2g44190.1 [Arabidopsis
thaliana] gi|7486486|pir||T00699 hypothetical protein
At2g44190 [imported] - Arabidopsis thaliana
gi|3212872|gb|AAC23423.1| hypothetical protein
[Arabidopsis thaliana]
Length = 474
Score = 177 bits (449), Expect = 2e-43
Identities = 90/188 (47%), Positives = 132/188 (69%)
Frame = -3
Query: 923 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 744
LQWRFANA A ++Q+ ++E+ YS +K+SE+ DSV RKR EL+ LQR K ++ I++
Sbjct: 282 LQWRFANANAEVKTQSQKAQAERMFYSLGLKMSELSDSVQRKRIELQHLQRVKAVTEIVE 341
Query: 743 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 564
+Q P LE+W+ +E+++S SL E T+AL+NAS++LP+ ++V+ +E+ EAL ASK ME
Sbjct: 342 SQTPSLEQWAVLEDEFSTSLLETTEALLNASLRLPLDSKIKVETKELAEALVVASKSMEG 401
Query: 563 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQVEECSLRGQLIQL 384
IV NI LVPK +E ++ +SELARV G E+A + + L KT+ SQ+EEC LR QLIQ
Sbjct: 402 IVQNIGNLVPKTQEMETLMSELARVSGIEKASVEDCRVALLKTHSSQMEECYLRSQLIQH 461
Query: 383 HSICQKNK 360
C + +
Sbjct: 462 QKKCHQQE 469
>ref|NP_191559.1| putative protein; protein id: At3g60000.1 [Arabidopsis thaliana]
gi|11282354|pir||T47824 hypothetical protein F24G16.270
- Arabidopsis thaliana gi|7019694|emb|CAB75819.1|
putative protein [Arabidopsis thaliana]
Length = 451
Score = 164 bits (414), Expect = 2e-39
Identities = 82/179 (45%), Positives = 121/179 (66%)
Frame = -3
Query: 923 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 744
LQWRFANA A + + ++E ++S KISE+ DSV RKR EL+ L + K L I +
Sbjct: 263 LQWRFANANAQVKTQTHKTQTETMIHSFGSKISELHDSVQRKRIELQRLLKTKALLAITE 322
Query: 743 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 564
+Q P LE+WS++EE+YS S+++ QA NAS++LP+ G++ VD +++G+ L +ASK+++
Sbjct: 323 SQTPCLEQWSAIEEEYSTSVSQTIQAFSNASLRLPLDGDIMVDSKQLGDGLVAASKIVDG 382
Query: 563 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQVEECSLRGQLIQ 387
I N+ +PKA+E +S +SEL RV ER+L L KT SQ+EECS+R QLIQ
Sbjct: 383 ITQNVGNYMPKAKEMESLLSELTRVARSERSLTENCVVALLKTQASQIEECSMRSQLIQ 441
>dbj|BAC42908.1| unknown protein [Arabidopsis thaliana] gi|28973259|gb|AAO63954.1|
unknown protein [Arabidopsis thaliana]
Length = 644
Score = 133 bits (335), Expect = 3e-30
Identities = 71/180 (39%), Positives = 118/180 (65%)
Frame = -3
Query: 923 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 744
LQWRFANA+A +++ Q+ +EK L++ + ISE+R SV KR +L L+++ L++IL
Sbjct: 462 LQWRFANARADSTLMVQRLSAEKILWNAWVSISELRHSVTLKRIKLLLMRQKLKLASILK 521
Query: 743 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 564
QM YLEEWS ++ ++S SL+ AT+AL ++++LPV G VD +++ A++SA +M
Sbjct: 522 EQMCYLEEWSLLDRNHSNSLSGATEALKASTLRLPVSGKAVVDIQDLKHAVSSAVDVMHA 581
Query: 563 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQVEECSLRGQLIQL 384
+VS+I L K EE +S ++E+ + G E L+ + L++ QV +CS++ +IQL
Sbjct: 582 MVSSIFSLTSKVEEMNSVMAEMVNITGKEEVLLEQCQGFLTRVAAMQVTDCSMKTHIIQL 641
>ref|NP_564558.1| expressed protein; protein id: At1g49890.1, supported by cDNA:
gi_15028144 [Arabidopsis thaliana]
gi|15028145|gb|AAK76696.1| unknown protein [Arabidopsis
thaliana] gi|24030506|gb|AAN41399.1| unknown protein
[Arabidopsis thaliana]
Length = 659
Score = 125 bits (313), Expect = 1e-27
Identities = 69/183 (37%), Positives = 115/183 (62%)
Frame = -3
Query: 923 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 744
LQWRF NA+A +++ Q+ +EK L++ + ISE+R SV KR +L LL++ L++IL
Sbjct: 467 LQWRFVNARADSTVMVQRLNAEKNLWNAWVSISELRHSVTLKRIKLLLLRQKLKLASILR 526
Query: 743 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 564
QM +LEEWS ++ D+S SL+ AT++L ++++LP+ G VD +++ A++SA +M+
Sbjct: 527 GQMGFLEEWSLLDRDHSSSLSGATESLKASTLRLPIVGKTVVDIQDLKHAVSSAVDVMQA 586
Query: 563 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQVEECSLRGQLIQL 384
+ S+I L K +E +S + E V E+ L+ LS+ QV +CS++ +IQL
Sbjct: 587 MSSSIFSLTSKVDEMNSVMVETVNVTAKEKVLLERCQGCLSRVAAMQVTDCSMKTHIIQL 646
Query: 383 HSI 375
I
Sbjct: 647 SRI 649
>pir||T52384 hypothetical protein MMB12.3 [imported] - Arabidopsis thaliana
gi|9294421|dbj|BAB02541.1|
emb|CAB52444.1~gene_id:MMB12.3~similar to unknown
protein [Arabidopsis thaliana]
Length = 701
Score = 120 bits (300), Expect = 4e-26
Identities = 65/167 (38%), Positives = 108/167 (63%)
Frame = -3
Query: 923 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 744
LQWRFANA+A +++ Q+ +EK L++ + ISE+R SV KR +L L+++ L++IL
Sbjct: 462 LQWRFANARADSTLMVQRLSAEKILWNAWVSISELRHSVTLKRIKLLLMRQKLKLASILK 521
Query: 743 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 564
QM YLEEWS ++ ++S SL+ AT+AL ++++LPV G VD +++ A++SA +M
Sbjct: 522 EQMCYLEEWSLLDRNHSNSLSGATEALKASTLRLPVSGKAVVDIQDLKHAVSSAVDVMHA 581
Query: 563 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQ 423
+VS+I L K EE +S ++E+ + G E L+ + L++ Q
Sbjct: 582 MVSSIFSLTSKVEEMNSVMAEMVNITGKEEVLLEQCQGFLTRVAAMQ 628
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 729,716,285
Number of Sequences: 1393205
Number of extensions: 15194651
Number of successful extensions: 39650
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 37606
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39575
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51027805888
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)