Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019976A_C01 KMC019976A_c01
(539 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
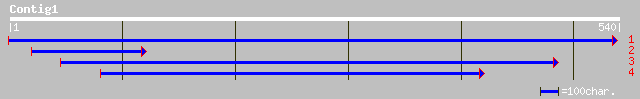
gb|AAK85402.1| beta-1,3-glucanase [Camellia sinensis] 126 1e-28
ref|NP_565269.1| glycosyl hydrolase family 17 (beta-1,3-glucanas... 109 2e-23
gb|AAN05325.1| Putative beta-1,3-glucanase [Oryza sativa (japoni... 107 7e-23
gb|AAO42272.1| unknown protein [Arabidopsis thaliana] 91 1e-17
ref|NP_172647.1| glycosyl hydrolase family 17; protein id: At1g1... 91 1e-17
>gb|AAK85402.1| beta-1,3-glucanase [Camellia sinensis]
Length = 350
Score = 126 bits (317), Expect = 1e-28
Identities = 61/110 (55%), Positives = 76/110 (68%)
Frame = -2
Query: 538 SSLLQGQPCYEPDNVISHASYAFDSYYHKMGNTPDSCDFKGVATITTSDPSHGSCKFPGS 359
S+LLQGQPCYEPD V +HA+YAFD+YY +MG +CDF GVATITT++P+HGSC FPGS
Sbjct: 242 SALLQGQPCYEPDTVAAHATYAFDTYYQQMGKASGTCDFNGVATITTTNPTHGSCVFPGS 301
Query: 358 LGKNGTFGNFTAPSMNSSSDSSAYNLQSCEMRARNLLMVTGFLLMGLVLM 209
GKNGTF N TAPS+NS+S A + ++ G L LV +
Sbjct: 302 NGKNGTFLNSTAPSLNSTSSGCPARYMRGSASA-SAIVAVGILSWSLVFL 350
>ref|NP_565269.1| glycosyl hydrolase family 17 (beta-1,3-glucanase); protein id:
At2g01630.1, supported by cDNA: 154469. [Arabidopsis
thaliana] gi|20197543|gb|AAD12708.2| putative
beta-1,3-glucanase [Arabidopsis thaliana]
gi|21553631|gb|AAM62724.1| putative beta-1,3-glucanase
[Arabidopsis thaliana]
Length = 501
Score = 109 bits (272), Expect = 2e-23
Identities = 49/82 (59%), Positives = 64/82 (77%), Gaps = 3/82 (3%)
Frame = -2
Query: 538 SSLLQGQPCYEPDNVISHASYAFDSYYHKMGNTPDSCDFKGVATITTSDPSHGSCKFPGS 359
S+L+QG+ CYEPD+V++H++YAF++YY KMG SCDFKGVAT+TT+DPS G+C FPGS
Sbjct: 388 SALMQGESCYEPDDVVAHSTYAFNAYYQKMGKASGSCDFKGVATVTTTDPSRGTCVFPGS 447
Query: 358 LGKNGTFGNFT---APSMNSSS 302
N T GN T APS NS++
Sbjct: 448 AKSNQTLGNNTSALAPSANSTT 469
>gb|AAN05325.1| Putative beta-1,3-glucanase [Oryza sativa (japonica
cultivar-group)]
Length = 504
Score = 107 bits (268), Expect = 7e-23
Identities = 55/113 (48%), Positives = 75/113 (65%), Gaps = 3/113 (2%)
Frame = -2
Query: 538 SSLLQGQPCYEPDNVISHASYAFDSYYHKMGNTPDSCDFKGVATITTSDPSHGSCKFPGS 359
S+L+QGQPCY+PDNV +HA+YAF++YYH MG +C F GVA ITT+DPSHGSC + GS
Sbjct: 392 SALMQGQPCYDPDNVEAHATYAFNAYYHGMGMGSGTCYFSGVAVITTTDPSHGSCVYAGS 451
Query: 358 LGKNGTF---GNFTAPSMNSSSDSSAYNLQSCEMRARNLLMVTGFLLMGLVLM 209
GKNGT G APS NS++ S + ++ + +V LL ++L+
Sbjct: 452 GGKNGTSLLNGTSLAPSSNSTAGDSGAHRAIGDVSSFVRAVVAALLLSVVLLL 504
>gb|AAO42272.1| unknown protein [Arabidopsis thaliana]
Length = 332
Score = 90.5 bits (223), Expect = 1e-17
Identities = 44/78 (56%), Positives = 53/78 (67%)
Frame = -2
Query: 538 SSLLQGQPCYEPDNVISHASYAFDSYYHKMGNTPDSCDFKGVATITTSDPSHGSCKFPGS 359
S + G+ CY+P+NV HAS+AF+SYY K G SCDFKGVA ITT+DPSHGSC FPGS
Sbjct: 230 SEIQPGESCYQPNNVKGHASFAFNSYYQKEGRASGSCDFKGVAMITTTDPSHGSCIFPGS 289
Query: 358 LGKNGTFGNFTAPSMNSS 305
GN T +NS+
Sbjct: 290 ----KKVGNRTQTVVNST 303
>ref|NP_172647.1| glycosyl hydrolase family 17; protein id: At1g11820.1 [Arabidopsis
thaliana]
Length = 511
Score = 90.5 bits (223), Expect = 1e-17
Identities = 44/78 (56%), Positives = 53/78 (67%)
Frame = -2
Query: 538 SSLLQGQPCYEPDNVISHASYAFDSYYHKMGNTPDSCDFKGVATITTSDPSHGSCKFPGS 359
S + G+ CY+P+NV HAS+AF+SYY K G SCDFKGVA ITT+DPSHGSC FPGS
Sbjct: 409 SEIQPGESCYQPNNVKGHASFAFNSYYQKEGRASGSCDFKGVAMITTTDPSHGSCIFPGS 468
Query: 358 LGKNGTFGNFTAPSMNSS 305
GN T +NS+
Sbjct: 469 ----KKVGNRTQTVVNST 482
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,329,877
Number of Sequences: 1393205
Number of extensions: 9812097
Number of successful extensions: 22824
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 22076
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22797
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)