Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019703A_C01 KMC019703A_c01
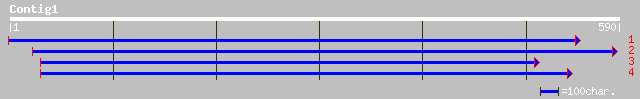
(589 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567971.1| chalcone synthase - like protein; protein id: A... 309 2e-83
pir||T10231 anther-specific protein homolog T11I11.90 - Arabidop... 303 1e-81
dbj|BAC21541.1| putative chalcone synthase [Oryza sativa (japoni... 268 4e-71
pir||T10742 chalcone synthase homolog ChS1 - Monterey pine gi|25... 255 3e-67
ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabi... 248 3e-65
>ref|NP_567971.1| chalcone synthase - like protein; protein id: At4g34850.1,
supported by cDNA: 19658. [Arabidopsis thaliana]
gi|21554049|gb|AAM63130.1| chalcone synthase-like
protein [Arabidopsis thaliana]
Length = 392
Score = 309 bits (791), Expect = 2e-83
Identities = 150/177 (84%), Positives = 164/177 (91%)
Frame = +3
Query: 57 GVTKQANPGKATILALGKAFPHQLVMQEYLVDGYFRDTNCDNPELKQKLTRLSKTTTVRT 236
G K++NPGKATILALGKAFPHQLVMQEYLVDGYF+ T CD+PELKQKLTRL KTTTV+T
Sbjct: 10 GSEKKSNPGKATILALGKAFPHQLVMQEYLVDGYFKTTKCDDPELKQKLTRLCKTTTVKT 69
Query: 237 RYVVMSEEILKKYPELAVEGTPTVKQRLEICNKAVTEMAIEASQVCIKSWGRALSDITHV 416
RYVVMSEEILKKYPELA+EG TV QRL+ICN AVTEMA+EAS+ CIK+WGR++SDITHV
Sbjct: 70 RYVVMSEEILKKYPELAIEGGSTVTQRLDICNDAVTEMAVEASRACIKNWGRSISDITHV 129
Query: 417 VYVSSSEARLPGGDLYLAKGLGLNPDTKRTMLYFAGCSGGVAGLRVAKDIAENNPGS 587
VYVSSSEARLPGGDLYLAKGLGL+PDT R +LYF GCSGGVAGLRVAKDIAENNPGS
Sbjct: 130 VYVSSSEARLPGGDLYLAKGLGLSPDTHRVLLYFVGCSGGVAGLRVAKDIAENNPGS 186
>pir||T10231 anther-specific protein homolog T11I11.90 - Arabidopsis thaliana
gi|5123702|emb|CAB45446.1| chalcone synthase-like
protein [Arabidopsis thaliana]
gi|7270436|emb|CAB80202.1| chalcone synthase-like
protein [Arabidopsis thaliana]
Length = 390
Score = 303 bits (775), Expect = 1e-81
Identities = 149/177 (84%), Positives = 163/177 (91%)
Frame = +3
Query: 57 GVTKQANPGKATILALGKAFPHQLVMQEYLVDGYFRDTNCDNPELKQKLTRLSKTTTVRT 236
G K++NPGKATILALGKAFPHQLVMQEYLVDGYF+ T CD+PELKQKLTR KTTTV+T
Sbjct: 10 GSEKKSNPGKATILALGKAFPHQLVMQEYLVDGYFKTTKCDDPELKQKLTR--KTTTVKT 67
Query: 237 RYVVMSEEILKKYPELAVEGTPTVKQRLEICNKAVTEMAIEASQVCIKSWGRALSDITHV 416
RYVVMSEEILKKYPELA+EG TV QRL+ICN AVTEMA+EAS+ CIK+WGR++SDITHV
Sbjct: 68 RYVVMSEEILKKYPELAIEGGSTVTQRLDICNDAVTEMAVEASRACIKNWGRSISDITHV 127
Query: 417 VYVSSSEARLPGGDLYLAKGLGLNPDTKRTMLYFAGCSGGVAGLRVAKDIAENNPGS 587
VYVSSSEARLPGGDLYLAKGLGL+PDT R +LYF GCSGGVAGLRVAKDIAENNPGS
Sbjct: 128 VYVSSSEARLPGGDLYLAKGLGLSPDTHRVLLYFVGCSGGVAGLRVAKDIAENNPGS 184
>dbj|BAC21541.1| putative chalcone synthase [Oryza sativa (japonica cultivar-group)]
Length = 405
Score = 268 bits (685), Expect = 4e-71
Identities = 130/171 (76%), Positives = 151/171 (88%)
Frame = +3
Query: 75 NPGKATILALGKAFPHQLVMQEYLVDGYFRDTNCDNPELKQKLTRLSKTTTVRTRYVVMS 254
NPGKATILALG AFP QLVMQ+Y+VDG+ R+TNCD+PELK+KLTRL KTTTV+TRYVVMS
Sbjct: 20 NPGKATILALGHAFPQQLVMQDYVVDGFMRNTNCDDPELKEKLTRLCKTTTVKTRYVVMS 79
Query: 255 EEILKKYPELAVEGTPTVKQRLEICNKAVTEMAIEASQVCIKSWGRALSDITHVVYVSSS 434
EEILK YPELA EG PT+KQRL+I NKAVT+MA EAS C++SWG ALS+ITH+VYVSSS
Sbjct: 80 EEILKSYPELAQEGQPTMKQRLDISNKAVTQMATEASLACVRSWGGALSEITHLVYVSSS 139
Query: 435 EARLPGGDLYLAKGLGLNPDTKRTMLYFAGCSGGVAGLRVAKDIAENNPGS 587
EAR PGGDL+LA+ LGL+PD +R ML F GCSGGVAGLRVAK +AE+ PG+
Sbjct: 140 EARFPGGDLHLARALGLSPDVRRVMLAFTGCSGGVAGLRVAKGLAESCPGA 190
>pir||T10742 chalcone synthase homolog ChS1 - Monterey pine
gi|2507617|gb|AAB80804.1| chalcone synthase homolog
PrChS1 [Pinus radiata]
Length = 390
Score = 255 bits (651), Expect = 3e-67
Identities = 123/174 (70%), Positives = 145/174 (82%)
Frame = +3
Query: 66 KQANPGKATILALGKAFPHQLVMQEYLVDGYFRDTNCDNPELKQKLTRLSKTTTVRTRYV 245
+Q PGK T +A G+AFP QLVMQE+LVDGYFR+TNC +P L+QKL RL KTTTV+TRYV
Sbjct: 17 RQHRPGKTTAMAFGRAFPDQLVMQEFLVDGYFRNTNCQDPVLRQKLERLCKTTTVKTRYV 76
Query: 246 VMSEEILKKYPELAVEGTPTVKQRLEICNKAVTEMAIEASQVCIKSWGRALSDITHVVYV 425
VMS+EIL ++PELAVEG+ TV+QRLEI N AVT+MA++A + C+K WGR +S+ITH+VYV
Sbjct: 77 VMSDEILAQHPELAVEGSATVRQRLEISNVAVTDMAVDACRDCLKEWGRPVSEITHLVYV 136
Query: 426 SSSEARLPGGDLYLAKGLGLNPDTKRTMLYFAGCSGGVAGLRVAKDIAENNPGS 587
SSSE RLPGGDLYLA LGL D R MLYF GC GGV GLRVAKD+AENNPGS
Sbjct: 137 SSSEIRLPGGDLYLASRLGLRSDVSRVMLYFLGCYGGVTGLRVAKDLAENNPGS 190
>ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabidopsis thaliana]
gi|25286664|pir||E86152 chalcone synthase homolog
T7I23.4 - Arabidopsis thaliana gi|2317904|gb|AAC24368.1|
Similar to rice chalcone synthase homolog,
gp|U90341|2507617 and anther specific protein,
gp|Y14507|2326772 [Arabidopsis thaliana]
Length = 395
Score = 248 bits (634), Expect = 3e-65
Identities = 122/178 (68%), Positives = 140/178 (78%)
Frame = +3
Query: 54 KGVTKQANPGKATILALGKAFPHQLVMQEYLVDGYFRDTNCDNPELKQKLTRLSKTTTVR 233
K + AN GKAT+LALGKAFP Q+V QE LV+G+ RDT CD+ +K+KL L KTTTV+
Sbjct: 15 KSTRRVANAGKATLLALGKAFPSQVVPQENLVEGFLRDTKCDDAFIKEKLEHLCKTTTVK 74
Query: 234 TRYVVMSEEILKKYPELAVEGTPTVKQRLEICNKAVTEMAIEASQVCIKSWGRALSDITH 413
TRY V++ EIL KYPEL EG+PT+KQRLEI N+AV EMA+EAS CIK WGR + DITH
Sbjct: 75 TRYTVLTREILAKYPELTTEGSPTIKQRLEIANEAVVEMALEASLGCIKEWGRPVEDITH 134
Query: 414 VVYVSSSEARLPGGDLYLAKGLGLNPDTKRTMLYFAGCSGGVAGLRVAKDIAENNPGS 587
+VYVSSSE RLPGGDLYL+ LGL D R MLYF GC GGV GLRVAKDIAENNPGS
Sbjct: 135 IVYVSSSEIRLPGGDLYLSAKLGLRNDVNRVMLYFLGCYGGVTGLRVAKDIAENNPGS 192
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,142,701
Number of Sequences: 1393205
Number of extensions: 11447843
Number of successful extensions: 32348
Number of sequences better than 10.0: 471
Number of HSP's better than 10.0 without gapping: 31182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32022
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)