Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019533A_C01 KMC019533A_c01
(585 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
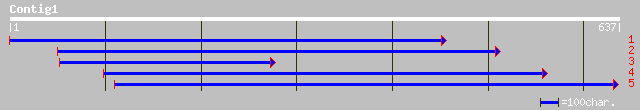
gb|AAG21908.1|AC026815_12 putative CER1 [Oryza sativa] 164 6e-40
ref|NP_181306.1| CER1-like protein; protein id: At2g37700.1 [Ara... 159 2e-38
gb|AAB87721.1| maize gl1 homolog [Arabidopsis thaliana] 155 4e-37
ref|NP_171721.1| hypothetical protein; protein id: At1g02190.1 [... 155 5e-37
gb|AAC24373.1| CER1-like protein [Arabidopsis thaliana] 155 5e-37
>gb|AAG21908.1|AC026815_12 putative CER1 [Oryza sativa]
Length = 621
Score = 164 bits (416), Expect = 6e-40
Identities = 71/115 (61%), Positives = 89/115 (76%), Gaps = 1/115 (0%)
Frame = -1
Query: 585 KSYNQTIWLVGDGLTEEEQVNAPXGTIFIPYSQFPPKKYRKD-CSYHCTPAMQIPSSVEN 409
K+ +WL+GDGL EQ A GT+FIPYSQFPPK RKD CSY TPAM +P +++N
Sbjct: 504 KTGTAKVWLIGDGLDSAEQFRAQKGTLFIPYSQFPPKMVRKDSCSYSTTPAMAVPKTLQN 563
Query: 408 IHSCEDWLPRRVMSAWRIAGIVHSLEGWTEHECGYTMHNIDKVWKSTLQHGFKPL 244
+HSCE+WLPRRVMSAWRIAGI+H+LEGW EHECG + ++DKVW + + HGF P+
Sbjct: 564 VHSCENWLPRRVMSAWRIAGILHALEGWNEHECGDKVLDMDKVWSAAIMHGFCPV 618
>ref|NP_181306.1| CER1-like protein; protein id: At2g37700.1 [Arabidopsis thaliana]
gi|7485434|pir||T02536 CER1-like protein [imported] -
Arabidopsis thaliana gi|3236252|gb|AAC23640.1| CER1-like
protein [Arabidopsis thaliana]
Length = 635
Score = 159 bits (403), Expect = 2e-38
Identities = 73/122 (59%), Positives = 92/122 (74%), Gaps = 3/122 (2%)
Frame = -1
Query: 582 SYNQTIWLVGDGLTEEEQVNAPXGTIFIPYSQFPPKKYRKDCSYHCTPAMQIPSSVENIH 403
+Y IWLVGDGL+ +EQ A GT+F+P+SQFPPK RKDC YH TPAM IP S +NI
Sbjct: 513 NYYPMIWLVGDGLSTKEQKMAKDGTLFLPFSQFPPKTLRKDCFYHTTPAMIIPHSAQNID 572
Query: 402 SCEDWLPRRVMSAWRIAGIVHSLEGWTEHECGY---TMHNIDKVWKSTLQHGFKPLTVPV 232
SCE+WL RRVMSAWR+ GIVH+LEGW EHECG ++ N +VW++ L++GF+PL +P
Sbjct: 573 SCENWLGRRVMSAWRVGGIVHALEGWKEHECGLDDNSIINPPRVWEAALRNGFQPLLLPS 632
Query: 231 QE 226
E
Sbjct: 633 LE 634
>gb|AAB87721.1| maize gl1 homolog [Arabidopsis thaliana]
Length = 625
Score = 155 bits (392), Expect = 4e-37
Identities = 66/113 (58%), Positives = 88/113 (77%), Gaps = 2/113 (1%)
Frame = -1
Query: 567 IWLVGDGLTEEEQVNAPXGTIFIPYSQFPPKKYRKDCSYHCTPAMQIPSSVENIHSCEDW 388
+WLVG+G T EEQ A GT+FIP+SQFP K+ R DC YH TPA+ +P S+ N+HSCE+W
Sbjct: 511 VWLVGEGTTREEQEKATKGTLFIPFSQFPLKQLRSDCIYHTTPALIVPKSLVNVHSCENW 570
Query: 387 LPRRVMSAWRIAGIVHSLEGWTEHECGYT--MHNIDKVWKSTLQHGFKPLTVP 235
LPR+ MSA R+AGI+H+LEGW HECG + + ++DKVW++ L HGF+PL +P
Sbjct: 571 LPRKAMSATRVAGILHALEGWETHECGTSLLLSDLDKVWEACLSHGFQPLLLP 623
>ref|NP_171721.1| hypothetical protein; protein id: At1g02190.1 [Arabidopsis
thaliana]
Length = 621
Score = 155 bits (391), Expect = 5e-37
Identities = 65/114 (57%), Positives = 86/114 (75%), Gaps = 2/114 (1%)
Frame = -1
Query: 579 YNQTIWLVGDGLTEEEQVNAPXGTIFIPYSQFPPKKYRKDCSYHCTPAMQIPSSVENIHS 400
Y+ +WLVGDG+ EEQ+ A GT+F+P+S FPP K RKDC Y TPAM++P S +NI S
Sbjct: 503 YSPKVWLVGDGIENEEQMKAKEGTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDS 562
Query: 399 CEDWLPRRVMSAWRIAGIVHSLEGWTEHECGYTMH--NIDKVWKSTLQHGFKPL 244
CE+WL RRVMSAW+I GIVH+LEGW EH+CG T + + +W++ L+H F+PL
Sbjct: 563 CENWLGRRVMSAWKIGGIVHALEGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 616
>gb|AAC24373.1| CER1-like protein [Arabidopsis thaliana]
Length = 604
Score = 155 bits (391), Expect = 5e-37
Identities = 65/114 (57%), Positives = 86/114 (75%), Gaps = 2/114 (1%)
Frame = -1
Query: 579 YNQTIWLVGDGLTEEEQVNAPXGTIFIPYSQFPPKKYRKDCSYHCTPAMQIPSSVENIHS 400
Y+ +WLVGDG+ EEQ+ A GT+F+P+S FPP K RKDC Y TPAM++P S +NI S
Sbjct: 486 YSPKVWLVGDGIENEEQMKAKEGTLFVPFSHFPPNKLRKDCFYQSTPAMRVPKSAQNIDS 545
Query: 399 CEDWLPRRVMSAWRIAGIVHSLEGWTEHECGYTMH--NIDKVWKSTLQHGFKPL 244
CE+WL RRVMSAW+I GIVH+LEGW EH+CG T + + +W++ L+H F+PL
Sbjct: 546 CENWLGRRVMSAWKIGGIVHALEGWEEHDCGNTCNVLRLHAIWEAALRHDFQPL 599
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 530,887,739
Number of Sequences: 1393205
Number of extensions: 11824413
Number of successful extensions: 29822
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 28945
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29793
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)