Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019238A_C01 KMC019238A_c01
(723 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
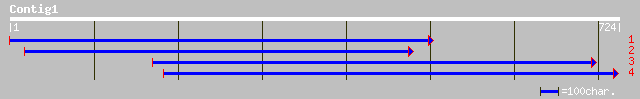
ref|NP_187728.1| putative leucoanthocyanidin dioxygenase; protei... 220 1e-56
ref|NP_196179.1| leucoanthocyanidin dioxygenase-like protein; pr... 215 4e-55
gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arab... 215 4e-55
gb|AAD52015.1|AF082862_1 unknown [Pisum sativum] 203 2e-51
ref|NP_191156.1| leucoanthocyanidin dioxygenase -like protein; p... 198 6e-50
>ref|NP_187728.1| putative leucoanthocyanidin dioxygenase; protein id: At3g11180.1
[Arabidopsis thaliana]
gi|6016680|gb|AAF01507.1|AC009991_3 putative
leucoanthocyanidin dioxygenase [Arabidopsis thaliana]
gi|12321884|gb|AAG50980.1|AC073395_22 leucoanthocyanidin
dioxygenase, putative; 41415-43854 [Arabidopsis
thaliana]
Length = 400
Score = 220 bits (561), Expect = 1e-56
Identities = 108/135 (80%), Positives = 118/135 (87%)
Frame = -2
Query: 722 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIY 543
L LGLS HSDPGGMT+LLPDDQV GLQVR GD WITVNP RHAFIVNIGDQIQ+LSN+ Y
Sbjct: 266 LALGLSPHSDPGGMTILLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKY 325
Query: 542 TSVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRMR 363
SVEHRVIVNS+KERVSLAFFYNPKSDIPI+P+++LVT P LYP MTFD+YRLFIR +
Sbjct: 326 KSVEHRVIVNSEKERVSLAFFYNPKSDIPIQPMQQLVTSTMPPLYPPMTFDQYRLFIRTQ 385
Query: 362 GPRGKSQVESMKSPR 318
GPRGKS VES SPR
Sbjct: 386 GPRGKSHVESHISPR 400
>ref|NP_196179.1| leucoanthocyanidin dioxygenase-like protein; protein id:
At5g05600.1, supported by cDNA: 13012., supported by
cDNA: gi_14532537 [Arabidopsis thaliana]
gi|10178137|dbj|BAB11549.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
gi|14532538|gb|AAK63997.1| AT5g05600/MOP10_14
[Arabidopsis thaliana] gi|22137300|gb|AAM91495.1|
AT5g05600/MOP10_14 [Arabidopsis thaliana]
Length = 371
Score = 215 bits (548), Expect = 4e-55
Identities = 106/135 (78%), Positives = 116/135 (85%)
Frame = -2
Query: 722 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIY 543
L LGLS HSDPGGMT+LLPDDQV GLQVRK D WITV P HAFIVNIGDQIQ+LSN+ Y
Sbjct: 237 LALGLSPHSDPGGMTILLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTY 296
Query: 542 TSVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRMR 363
SVEHRVIVNSDKERVSLAFFYNPKSDIPI+P++ELV+ P LYP MTFD+YRLFIR +
Sbjct: 297 KSVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQ 356
Query: 362 GPRGKSQVESMKSPR 318
GP+GKS VES SPR
Sbjct: 357 GPQGKSHVESHISPR 371
>gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arabidopsis thaliana]
Length = 355
Score = 215 bits (548), Expect = 4e-55
Identities = 106/135 (78%), Positives = 116/135 (85%)
Frame = -2
Query: 722 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIY 543
L LGLS HSDPGGMT+LLPDDQV GLQVRK D WITV P HAFIVNIGDQIQ+LSN+ Y
Sbjct: 221 LALGLSPHSDPGGMTILLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTY 280
Query: 542 TSVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRMR 363
SVEHRVIVNSDKERVSLAFFYNPKSDIPI+P++ELV+ P LYP MTFD+YRLFIR +
Sbjct: 281 KSVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQ 340
Query: 362 GPRGKSQVESMKSPR 318
GP+GKS VES SPR
Sbjct: 341 GPQGKSHVESHISPR 355
>gb|AAD52015.1|AF082862_1 unknown [Pisum sativum]
Length = 134
Score = 203 bits (516), Expect = 2e-51
Identities = 96/131 (73%), Positives = 115/131 (87%)
Frame = -2
Query: 716 LGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIYTS 537
+GL+ H+DPGGMT+LLPDD V GLQVRKG+DWITV P +AFI+NIGDQIQV+SNAIY S
Sbjct: 2 MGLAPHTDPGGMTILLPDDFVSGLQVRKGNDWITVKPVPNAFIINIGDQIQVMSNAIYKS 61
Query: 536 VEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRMRGP 357
VEHRVIVN ++RVSLA FYNPKSD+ I+P KELVT ++PALYP MT+DEYRL+IRM+GP
Sbjct: 62 VEHRVIVNPTQDRVSLAMFYNPKSDLIIQPAKELVTKERPALYPPMTYDEYRLYIRMKGP 121
Query: 356 RGKSQVESMKS 324
GK+QVES+ S
Sbjct: 122 CGKAQVESLAS 132
>ref|NP_191156.1| leucoanthocyanidin dioxygenase -like protein; protein id:
At3g55970.1 [Arabidopsis thaliana]
gi|11255840|pir||T49209 leucoanthocyanidin
dioxygenase-like protein - Arabidopsis thaliana
gi|7573492|emb|CAB87851.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
gi|12043537|emb|CAC19787.1| putative leucoanthocyanidin
dioxygenase [Arabidopsis thaliana]
Length = 363
Score = 198 bits (504), Expect = 6e-50
Identities = 95/136 (69%), Positives = 113/136 (82%), Gaps = 1/136 (0%)
Frame = -2
Query: 722 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDD-WITVNPARHAFIVNIGDQIQVLSNAI 546
LTLG+S HSDPGG+T+LLPD+QV LQVR DD WITV PA HAFIVN+GDQIQ+LSN+I
Sbjct: 228 LTLGISPHSDPGGLTILLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSI 287
Query: 545 YTSVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRM 366
Y SVEHRVIVN + ER+SLAFFYNPK ++PIEP+KELVT D PALY + T+D YR FIR
Sbjct: 288 YKSVEHRVIVNPENERLSLAFFYNPKGNVPIEPLKELVTVDSPALYSSTTYDRYRQFIRT 347
Query: 365 RGPRGKSQVESMKSPR 318
+GPR K ++ +KSPR
Sbjct: 348 QGPRSKCHIDELKSPR 363
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 655,718,281
Number of Sequences: 1393205
Number of extensions: 14856844
Number of successful extensions: 35697
Number of sequences better than 10.0: 651
Number of HSP's better than 10.0 without gapping: 33893
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35355
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33780557640
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)