Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019168A_C01 KMC019168A_c01
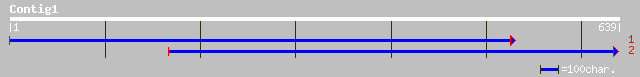
(639 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA36424.1| MYB homologue [Perilla frutescens] 53 4e-06
pir||T51638 probable transcription factor MYB17 [imported] - Ara... 49 4e-05
ref|NP_191684.1| myb DNA-binding protein (MYB17); protein id: At... 49 4e-05
gb|AAF80075.1|AF262187_1 immunoglobulin heavy chain variable reg... 33 2.4
dbj|BAC02317.1| immunoglobulin heavy chain VHDJ region [Homo sap... 32 5.5
>dbj|BAA36424.1| MYB homologue [Perilla frutescens]
Length = 282
Score = 52.8 bits (125), Expect = 4e-06
Identities = 26/37 (70%), Positives = 32/37 (86%)
Frame = -1
Query: 567 GSDSSCSNDLEDSSDTALQLLLDFPIYNDMSFLEGNS 457
G+DSS SN++EDSS++ LQLLLDFP NDMSFL GN+
Sbjct: 220 GTDSSSSNEMEDSSESTLQLLLDFPCKNDMSFL-GNA 255
>pir||T51638 probable transcription factor MYB17 [imported] - Arabidopsis
thaliana (fragment) gi|3941424|gb|AAC83588.1| putative
transcription factor [Arabidopsis thaliana]
Length = 224
Score = 49.3 bits (116), Expect = 4e-05
Identities = 32/47 (68%), Positives = 36/47 (76%), Gaps = 3/47 (6%)
Frame = -1
Query: 591 EEDVVQHHGSD-SSCSNDLED-SSDTALQLLLDFPIY-NDMSFLEGN 460
+E V HGSD S SNDLED S+D+ALQLLLDFPI +DMSFLE N
Sbjct: 159 KEVTVAIHGSDYSPYSNDLEDDSTDSALQLLLDFPISDDDMSFLEEN 205
>ref|NP_191684.1| myb DNA-binding protein (MYB17); protein id: At3g61250.1
[Arabidopsis thaliana] gi|11273969|pir||T47917 probable
transcription factor MYB17 - Arabidopsis thaliana
gi|6850892|emb|CAB71055.1| putative transcription factor
(MYB17) [Arabidopsis thaliana]
Length = 299
Score = 49.3 bits (116), Expect = 4e-05
Identities = 32/47 (68%), Positives = 36/47 (76%), Gaps = 3/47 (6%)
Frame = -1
Query: 591 EEDVVQHHGSD-SSCSNDLED-SSDTALQLLLDFPIY-NDMSFLEGN 460
+E V HGSD S SNDLED S+D+ALQLLLDFPI +DMSFLE N
Sbjct: 234 KEVTVAIHGSDYSPYSNDLEDDSTDSALQLLLDFPISDDDMSFLEEN 280
>gb|AAF80075.1|AF262187_1 immunoglobulin heavy chain variable region [Homo sapiens]
Length = 133
Score = 33.5 bits (75), Expect = 2.4
Identities = 21/68 (30%), Positives = 28/68 (40%)
Frame = +3
Query: 249 TLSRHTKGHGFKW*SAIPNAGSVNGF*SHRMGIRTAMDVALTNIGLQFHDVAATTTATAV 428
T R G G +W I +AGS S R + T++D A L V A TA
Sbjct: 35 TWIRQPPGKGLEWIGDISHAGSTINNPSLRSRVTTSLDAAKNQFSLVLTSVTAADTAVYY 94
Query: 429 SIPARGDK 452
+ GD+
Sbjct: 95 CVRKTGDR 102
>dbj|BAC02317.1| immunoglobulin heavy chain VHDJ region [Homo sapiens]
Length = 114
Score = 32.3 bits (72), Expect = 5.5
Identities = 21/68 (30%), Positives = 32/68 (46%)
Frame = +3
Query: 243 HTTLSRHTKGHGFKW*SAIPNAGSVNGF*SHRMGIRTAMDVALTNIGLQFHDVAATTTAT 422
H + R G G +W I ++GS N S + + +MD++ + L + V T T
Sbjct: 33 HWSWVRQPPGKGLEWIGEINHSGSTNYNSSLKSRVTMSMDMSKNQVSLNLNSV--TAADT 90
Query: 423 AVSIPARG 446
AV ARG
Sbjct: 91 AVYFCARG 98
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 523,534,429
Number of Sequences: 1393205
Number of extensions: 10536005
Number of successful extensions: 22497
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 21368
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22333
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26723359358
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)