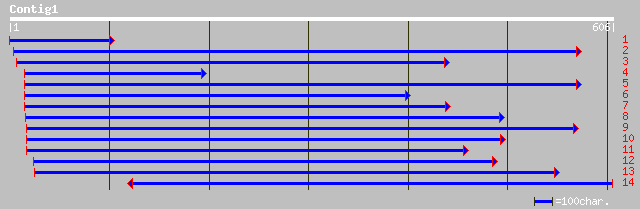
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019108A_C01 KMC019108A_c01
(568 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T02049 lipid transfer protein (clone ant43D) - common tobac... 93 3e-18
pir||T02048 lipid transfer protein (clone ant43C) - common tobac... 87 1e-16
gb|AAF65316.1|AF233297_1 lipid transfer protein [Nicotiana tabacum] 86 2e-16
gb|AAF71695.1|AF198168_1 phospholipid transfer protein [Aerides ... 81 1e-14
sp|P19656|NLTP_MAIZE Nonspecific lipid-transfer protein precurso... 80 2e-14
>pir||T02049 lipid transfer protein (clone ant43D) - common tobacco
gi|537639|gb|AAA21438.1| lipid transfer protein
Length = 118
Score = 92.8 bits (229), Expect = 3e-18
Identities = 43/113 (38%), Positives = 62/113 (54%)
Frame = -2
Query: 558 IFLAGLLPIVILLSISAAQATITCPAVIQAVTPCASFLLQNSSPSQGCCDGVKKLSGEAG 379
+FLA L +VI+ A A +CP V + PC S++ PS CC G+ + A
Sbjct: 6 VFLA--LALVIISKKGALGAPPSCPTVTTQLAPCLSYIQGGGDPSVPCCTGINNIYELAK 63
Query: 378 TQEGKTAICQCLKQGLAQVGKYDPNRVSELPKDCGVPLTLPPIDQNTDCTKVN 220
T+E + AIC CLK G +P V++LPK CG+ +PPID+N DC ++
Sbjct: 64 TKEDRVAICNCLKTAFTHAGNVNPTLVAQLPKKCGISFNMPPIDKNYDCNTIS 116
>pir||T02048 lipid transfer protein (clone ant43C) - common tobacco
gi|537637|gb|AAA21437.1| lipid transfer protein
Length = 120
Score = 87.4 bits (215), Expect = 1e-16
Identities = 45/116 (38%), Positives = 62/116 (52%), Gaps = 4/116 (3%)
Frame = -2
Query: 555 FLAGLLPIVILLSISAAQATITCPAVIQAVTPCASFLLQN----SSPSQGCCDGVKKLSG 388
FLA L +VI LS A A +C V + PC S++ +PS CC G+ +
Sbjct: 4 FLA-LALVVIALSNDALGAPPSCQTVTTQLAPCLSYIQNRVKGGGNPSVPCCTGINNIYE 62
Query: 387 EAGTQEGKTAICQCLKQGLAQVGKYDPNRVSELPKDCGVPLTLPPIDQNTDCTKVN 220
A T+E + AIC CLK G +P V+ELPK CG+ +PPID+N DC ++
Sbjct: 63 LAKTKEDRVAICNCLKNAFIHAGNVNPTLVAELPKKCGISFNMPPIDKNYDCNTIS 118
>gb|AAF65316.1|AF233297_1 lipid transfer protein [Nicotiana tabacum]
Length = 116
Score = 86.3 bits (212), Expect = 2e-16
Identities = 45/112 (40%), Positives = 60/112 (53%), Gaps = 4/112 (3%)
Frame = -2
Query: 555 FLAGLLPIVILLSISAAQATITCPAVIQAVTPCASFLLQN----SSPSQGCCDGVKKLSG 388
FLA L +VI LS A A +C V + PC S++ +PS CC G+ +
Sbjct: 4 FLA-LALVVIALSNDALGAPPSCQTVTTQLAPCLSYIQNRVKGGGNPSVPCCTGINNIYE 62
Query: 387 EAGTQEGKTAICQCLKQGLAQVGKYDPNRVSELPKDCGVPLTLPPIDQNTDC 232
A T+E + AIC CLK G +P V+ELPK CG+ +PPID+N DC
Sbjct: 63 LAKTKEDRVAICNCLKNAFIHAGNVNPTLVAELPKKCGISFNMPPIDKNYDC 114
>gb|AAF71695.1|AF198168_1 phospholipid transfer protein [Aerides japonica]
Length = 120
Score = 80.9 bits (198), Expect = 1e-14
Identities = 39/95 (41%), Positives = 55/95 (57%)
Frame = -2
Query: 507 AQATITCPAVIQAVTPCASFLLQNSSPSQGCCDGVKKLSGEAGTQEGKTAICQCLKQGLA 328
A TITC V+ +TPC S++ +S+ Q CC GVKKL+ A T + C CLK +
Sbjct: 26 ASGTITCGQVVSTLTPCISYIRGDSTLPQTCCSGVKKLNALASTSPDRQGACSCLKNLAS 85
Query: 327 QVGKYDPNRVSELPKDCGVPLTLPPIDQNTDCTKV 223
+ +P R + LP +CGV + PI +TDC+KV
Sbjct: 86 HIPNLNPARAAGLPGNCGVSVPY-PISTSTDCSKV 119
>sp|P19656|NLTP_MAIZE Nonspecific lipid-transfer protein precursor (LTP) (Phospholipid
transfer protein) (PLTP) (Allergen Zea m 14)
gi|82711|pir||A31779 phospholipid transfer protein 9C2
precursor - maize gi|168576|gb|AAA33493.1| phospholipid
transfer protein precursor
Length = 120
Score = 79.7 bits (195), Expect = 2e-14
Identities = 39/105 (37%), Positives = 58/105 (55%), Gaps = 1/105 (0%)
Frame = -2
Query: 531 VILLSISAAQATITCPAVIQAVTPCASFLL-QNSSPSQGCCDGVKKLSGEAGTQEGKTAI 355
++LL+ + ++A I+C V A+ PC S+ Q S PS GCC GV+ L+ A T + A
Sbjct: 17 LVLLAAATSEAAISCGQVASAIAPCISYARGQGSGPSAGCCSGVRSLNNAARTTADRRAA 76
Query: 354 CQCLKQGLAQVGKYDPNRVSELPKDCGVPLTLPPIDQNTDCTKVN 220
C CLK A V + + +P CGV + I +TDC++VN
Sbjct: 77 CNCLKNAAAGVSGLNAGNAASIPSKCGVSIPY-TISTSTDCSRVN 120
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 512,428,300
Number of Sequences: 1393205
Number of extensions: 11203204
Number of successful extensions: 28602
Number of sequences better than 10.0: 219
Number of HSP's better than 10.0 without gapping: 27419
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28496
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)