Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019105A_C01 KMC019105A_c01
(651 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
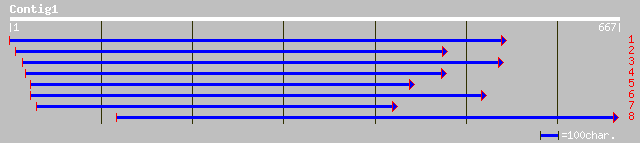
pir||S33804 male sterility protein 2 - Arabidopsis thaliana gi|3... 239 3e-62
ref|NP_187805.1| male sterility protein 2 (MS2); protein id: At3... 238 7e-62
pir||T08096 male sterility protein 2 - rape gi|1491638|emb|CAA68... 234 6e-61
gb|AAL84297.1|AC073556_14 putative male sterility protein [Oryza... 191 1e-47
ref|NP_191229.1| putative protein; protein id: At3g56700.1 [Arab... 102 4e-21
>pir||S33804 male sterility protein 2 - Arabidopsis thaliana
gi|396835|emb|CAA52019.1| male sterility 2 (MS2) protein
[Arabidopsis thaliana] gi|448297|prf||1916413A male
sterility 2 gene
Length = 616
Score = 239 bits (609), Expect = 3e-62
Identities = 112/155 (72%), Positives = 134/155 (86%), Gaps = 1/155 (0%)
Frame = -1
Query: 651 VASSVVNPLIFQDLARLLHEHYSSSPCFDSKGRPIQVPLMKLFSSTEEFSGHLWKDAIQK 472
+ASS +NPL+F+DLA LL+ HY +SPC DSKG PI V LMKLF+S ++FS HLW+DA ++
Sbjct: 461 IASSAINPLVFEDLAELLYNHYKTSPCMDSKGDPIMVRLMKLFNSVDDFSDHLWRDAQER 520
Query: 471 SGL-TAMASSKGKMSQKLENICRKSVEQAKYLANIYEPYTFYGGRFDNSNTQRLMESMSE 295
SGL + M+S+ KM QKL+ IC+KSVEQAK+LA IYEPYTFYGGRFDNSNTQRLME+MSE
Sbjct: 521 SGLMSGMSSADSKMMQKLKFICKKSVEQAKHLATIYEPYTFYGGRFDNSNTQRLMENMSE 580
Query: 294 DEKKEFGFDVKGIDWKDYITNVHIPGLRRHVMKGR 190
DEK+EFGFDV I+W DYITNVHIPGLRRHV+KGR
Sbjct: 581 DEKREFGFDVGSINWTDYITNVHIPGLRRHVLKGR 615
>ref|NP_187805.1| male sterility protein 2 (MS2); protein id: At3g11980.1
[Arabidopsis thaliana] gi|21431786|sp|Q08891|MS2_ARATH
Male sterility protein 2 gi|10998139|dbj|BAB03110.1|
male sterility protein 2 [Arabidopsis thaliana]
gi|12322016|gb|AAG51054.1|AC069473_16 male sterility
protein 2 (MS2); 67648-65205 [Arabidopsis thaliana]
Length = 616
Score = 238 bits (606), Expect = 7e-62
Identities = 112/155 (72%), Positives = 133/155 (85%), Gaps = 1/155 (0%)
Frame = -1
Query: 651 VASSVVNPLIFQDLARLLHEHYSSSPCFDSKGRPIQVPLMKLFSSTEEFSGHLWKDAIQK 472
+ASS +NPL+F+DLA LL+ HY +SPC DSKG PI V LMKLF+S ++FS HLW+DA ++
Sbjct: 461 IASSAINPLVFEDLAELLYNHYKTSPCMDSKGDPIMVRLMKLFNSVDDFSDHLWRDAQER 520
Query: 471 SGL-TAMASSKGKMSQKLENICRKSVEQAKYLANIYEPYTFYGGRFDNSNTQRLMESMSE 295
SGL + M+S KM QKL+ IC+KSVEQAK+LA IYEPYTFYGGRFDNSNTQRLME+MSE
Sbjct: 521 SGLMSGMSSVDSKMMQKLKFICKKSVEQAKHLATIYEPYTFYGGRFDNSNTQRLMENMSE 580
Query: 294 DEKKEFGFDVKGIDWKDYITNVHIPGLRRHVMKGR 190
DEK+EFGFDV I+W DYITNVHIPGLRRHV+KGR
Sbjct: 581 DEKREFGFDVGSINWTDYITNVHIPGLRRHVLKGR 615
>pir||T08096 male sterility protein 2 - rape gi|1491638|emb|CAA68190.1| male
sterility protein 2 [Brassica napus]
Length = 616
Score = 234 bits (598), Expect = 6e-61
Identities = 110/155 (70%), Positives = 131/155 (83%), Gaps = 1/155 (0%)
Frame = -1
Query: 651 VASSVVNPLIFQDLARLLHEHYSSSPCFDSKGRPIQVPLMKLFSSTEEFSGHLWKDAIQK 472
+ASS +NPL+F+DLA LL+ HY S+PC DSKG PI+VPLMKLF S ++FS HLW+DA ++
Sbjct: 461 IASSAINPLVFEDLAELLYNHYKSTPCMDSKGVPIRVPLMKLFDSVDDFSDHLWRDAQER 520
Query: 471 SGL-TAMASSKGKMSQKLENICRKSVEQAKYLANIYEPYTFYGGRFDNSNTQRLMESMSE 295
SGL M SS K+ QKL+ IC+KS+EQAK+LA IYEPYTFYGGRFDNSNT RLME+MSE
Sbjct: 521 SGLMNGMDSSDSKILQKLKFICKKSIEQAKHLATIYEPYTFYGGRFDNSNTHRLMENMSE 580
Query: 294 DEKKEFGFDVKGIDWKDYITNVHIPGLRRHVMKGR 190
+EK EFGFDV I+W DYITNVHIPGLRRHV+KGR
Sbjct: 581 EEKLEFGFDVGSINWNDYITNVHIPGLRRHVLKGR 615
>gb|AAL84297.1|AC073556_14 putative male sterility protein [Oryza sativa (japonica
cultivar-group)]
Length = 608
Score = 191 bits (484), Expect = 1e-47
Identities = 91/160 (56%), Positives = 119/160 (73%), Gaps = 3/160 (1%)
Frame = -1
Query: 651 VASSVVNPLIFQDLARLLHEHYSSSPCFDSKGRPIQVPLMKLFSSTEEFSGHLWKDAIQK 472
VASS VNPL F DL+R L +H++ SP D+ GRPI VP M+LF + E+F+ ++ DA+ +
Sbjct: 430 VASSTVNPLAFGDLSRFLFQHFTGSPYSDAAGRPIHVPPMRLFDTMEQFASYVETDALLR 489
Query: 471 SGLTAMASSKG---KMSQKLENICRKSVEQAKYLANIYEPYTFYGGRFDNSNTQRLMESM 301
+G A A + ++SQ+L +C KSVEQ YL +IY+PYTFYGGRFDN NT+ L+ M
Sbjct: 490 AGRLAGAGAGAGDERVSQRLRELCAKSVEQTIYLGSIYQPYTFYGGRFDNGNTEALIGEM 549
Query: 300 SEDEKKEFGFDVKGIDWKDYITNVHIPGLRRHVMKGRGMG 181
SE+EK F FDV+ I+W DYITNVHIPGLR+HVMKGRG+G
Sbjct: 550 SEEEKARFHFDVRSIEWTDYITNVHIPGLRKHVMKGRGVG 589
>ref|NP_191229.1| putative protein; protein id: At3g56700.1 [Arabidopsis thaliana]
gi|11358436|pir||T51258 hypothetical protein T8M16_30 -
Arabidopsis thaliana gi|9662989|emb|CAC00733.1| putative
protein [Arabidopsis thaliana]
Length = 527
Score = 102 bits (254), Expect = 4e-21
Identities = 59/150 (39%), Positives = 87/150 (57%)
Frame = -1
Query: 648 ASSVVNPLIFQDLARLLHEHYSSSPCFDSKGRPIQVPLMKLFSSTEEFSGHLWKDAIQKS 469
+SS VNP+ L L H+H P ++ I + MK+ SS E F+ L I++
Sbjct: 381 SSSHVNPMRAGKLIDLSHQHLCDFPLEETV---IDLEHMKIHSSLEGFTSALSNTIIKQE 437
Query: 468 GLTAMASSKGKMSQKLENICRKSVEQAKYLANIYEPYTFYGGRFDNSNTQRLMESMSEDE 289
+ + + G +S K ++ + LA YEPYTF+ RFDN+NT L++ MS +E
Sbjct: 438 RV--IDNEGGGLSTK----GKRKLNYFVSLAKTYEPYTFFQARFDNTNTTSLIQEMSMEE 491
Query: 288 KKEFGFDVKGIDWKDYITNVHIPGLRRHVM 199
KK FGFD+KGIDW+ YI NVH+PGL++ +
Sbjct: 492 KKTFGFDIKGIDWEHYIVNVHLPGLKKEFL 521
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 571,937,500
Number of Sequences: 1393205
Number of extensions: 12520596
Number of successful extensions: 35537
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 34478
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35511
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)