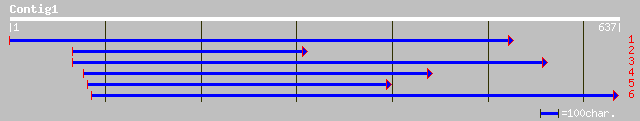
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019101A_C01 KMC019101A_c01
(634 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T10231 anther-specific protein homolog T11I11.90 - Arabidop... 221 5e-57
ref|NP_567971.1| chalcone synthase - like protein; protein id: A... 221 5e-57
pir||T10742 chalcone synthase homolog ChS1 - Monterey pine gi|25... 187 1e-46
ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabi... 184 6e-46
pir||S32145 anther-specific protein - rape (fragment) 182 3e-45
>pir||T10231 anther-specific protein homolog T11I11.90 - Arabidopsis thaliana
gi|5123702|emb|CAB45446.1| chalcone synthase-like
protein [Arabidopsis thaliana]
gi|7270436|emb|CAB80202.1| chalcone synthase-like
protein [Arabidopsis thaliana]
Length = 390
Score = 221 bits (564), Expect = 5e-57
Identities = 105/139 (75%), Positives = 122/139 (87%)
Frame = -2
Query: 633 TEKKIDGRLTEEGISFKLARELPQIIEDTVEGFCEKLMGVVGFQNKEYNNLFWAVHPGGP 454
TEK IDGRLTE+GI+FKL+RELPQIIED VE FC+KL+G G +K YN +FWAVHPGGP
Sbjct: 250 TEKTIDGRLTEQGINFKLSRELPQIIEDNVENFCKKLIGKAGLAHKNYNQMFWAVHPGGP 309
Query: 453 AILNRIEKKLDLLPEKLNASRRALMDFGNASSNTIVYVLEYMMEEGLKIRKGGEEDPEWG 274
AILNRIEK+L+L PEKL+ SRRALMD+GNASSN+IVYVLEYM+EE K+R EE+ EWG
Sbjct: 310 AILNRIEKRLNLSPEKLSPSRRALMDYGNASSNSIVYVLEYMLEESKKVRNMNEEENEWG 369
Query: 273 LILAFGPGITFEGILARNL 217
LILAFGPG+TFEGI+ARNL
Sbjct: 370 LILAFGPGVTFEGIIARNL 388
>ref|NP_567971.1| chalcone synthase - like protein; protein id: At4g34850.1,
supported by cDNA: 19658. [Arabidopsis thaliana]
gi|21554049|gb|AAM63130.1| chalcone synthase-like
protein [Arabidopsis thaliana]
Length = 392
Score = 221 bits (564), Expect = 5e-57
Identities = 105/139 (75%), Positives = 122/139 (87%)
Frame = -2
Query: 633 TEKKIDGRLTEEGISFKLARELPQIIEDTVEGFCEKLMGVVGFQNKEYNNLFWAVHPGGP 454
TEK IDGRLTE+GI+FKL+RELPQIIED VE FC+KL+G G +K YN +FWAVHPGGP
Sbjct: 252 TEKTIDGRLTEQGINFKLSRELPQIIEDNVENFCKKLIGKAGLAHKNYNQMFWAVHPGGP 311
Query: 453 AILNRIEKKLDLLPEKLNASRRALMDFGNASSNTIVYVLEYMMEEGLKIRKGGEEDPEWG 274
AILNRIEK+L+L PEKL+ SRRALMD+GNASSN+IVYVLEYM+EE K+R EE+ EWG
Sbjct: 312 AILNRIEKRLNLSPEKLSPSRRALMDYGNASSNSIVYVLEYMLEESKKVRNMNEEENEWG 371
Query: 273 LILAFGPGITFEGILARNL 217
LILAFGPG+TFEGI+ARNL
Sbjct: 372 LILAFGPGVTFEGIIARNL 390
>pir||T10742 chalcone synthase homolog ChS1 - Monterey pine
gi|2507617|gb|AAB80804.1| chalcone synthase homolog
PrChS1 [Pinus radiata]
Length = 390
Score = 187 bits (475), Expect = 1e-46
Identities = 96/139 (69%), Positives = 110/139 (79%)
Frame = -2
Query: 633 TEKKIDGRLTEEGISFKLARELPQIIEDTVEGFCEKLMGVVGFQNKEYNNLFWAVHPGGP 454
T I+GRLTEEGI+FKL RELPQIIED +EGFC KLM G +YN LFW VHPGGP
Sbjct: 257 THGTINGRLTEEGINFKLGRELPQIIEDHIEGFCRKLMDKAGVD--DYNELFWGVHPGGP 314
Query: 453 AILNRIEKKLDLLPEKLNASRRALMDFGNASSNTIVYVLEYMMEEGLKIRKGGEEDPEWG 274
AILNR+EKKL L PEKL SR+AL D+GNASSNTIVYVL+ M + ++ G ++ PEWG
Sbjct: 315 AILNRLEKKLSLGPEKLYYSRQALADYGNASSNTIVYVLDAMRQ----LKGGEKQSPEWG 370
Query: 273 LILAFGPGITFEGILARNL 217
LILAFGPGITFEGILAR+L
Sbjct: 371 LILAFGPGITFEGILARSL 389
>ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabidopsis thaliana]
gi|25286664|pir||E86152 chalcone synthase homolog
T7I23.4 - Arabidopsis thaliana gi|2317904|gb|AAC24368.1|
Similar to rice chalcone synthase homolog,
gp|U90341|2507617 and anther specific protein,
gp|Y14507|2326772 [Arabidopsis thaliana]
Length = 395
Score = 184 bits (468), Expect = 6e-46
Identities = 89/139 (64%), Positives = 113/139 (81%)
Frame = -2
Query: 633 TEKKIDGRLTEEGISFKLARELPQIIEDTVEGFCEKLMGVVGFQNKEYNNLFWAVHPGGP 454
T+ I+GRLTEEGI+FKL R+LPQ IE+ +E FC+KLMG G ++ E+N++FWAVHPGGP
Sbjct: 257 TQNVIEGRLTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDESMEFNDMFWAVHPGGP 316
Query: 453 AILNRIEKKLDLLPEKLNASRRALMDFGNASSNTIVYVLEYMMEEGLKIRKGGEEDPEWG 274
AILNR+E KL L EKL +SRRAL+D+GN SSNTI+YV+EYM +E ++K G+ EWG
Sbjct: 317 AILNRLETKLKLEKEKLESSRRALVDYGNVSSNTILYVMEYMRDE---LKKKGDAAQEWG 373
Query: 273 LILAFGPGITFEGILARNL 217
L LAFGPGITFEG+L R+L
Sbjct: 374 LGLAFGPGITFEGLLIRSL 392
>pir||S32145 anther-specific protein - rape (fragment)
Length = 190
Score = 182 bits (462), Expect = 3e-45
Identities = 89/141 (63%), Positives = 111/141 (78%)
Frame = -2
Query: 633 TEKKIDGRLTEEGISFKLARELPQIIEDTVEGFCEKLMGVVGFQNKEYNNLFWAVHPGGP 454
T+ IDGRLTEEGI+FKL R+LPQ IE+ +E FC+KLMG G + E+N++FW VHPGGP
Sbjct: 52 TQTVIDGRLTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDDSMEFNDMFWTVHPGGP 111
Query: 453 AILNRIEKKLDLLPEKLNASRRALMDFGNASSNTIVYVLEYMMEEGLKIRKGGEEDPEWG 274
AILNR+E KL L EKL SRRAL+D+GN SSNTI+YV+EYM +E ++K G+ EWG
Sbjct: 112 AILNRLETKLKLGREKLECSRRALVDYGNVSSNTILYVMEYMRDE---LKKKGDGAQEWG 168
Query: 273 LILAFGPGITFEGILARNLGA 211
L LAFGPGITFEG+L R+L +
Sbjct: 169 LGLAFGPGITFEGLLLRSLAS 189
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,458,959
Number of Sequences: 1393205
Number of extensions: 11695322
Number of successful extensions: 33732
Number of sequences better than 10.0: 531
Number of HSP's better than 10.0 without gapping: 31934
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33330
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)