Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019094A_C01 KMC019094A_c01
(1092 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
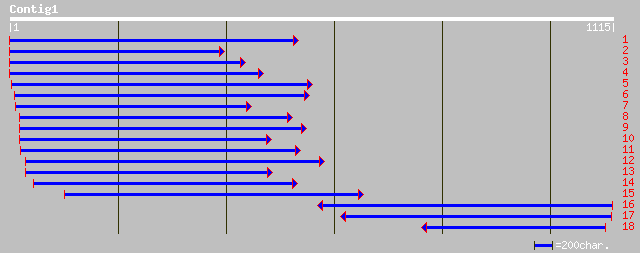
gb|AAN77692.1|AF487826_1 putative serine hydrolase [Vitis vinifera] 374 e-102
gb|AAD04946.2| PrMC3 [Pinus radiata] 303 2e-81
dbj|BAB44070.1| P0030H07.40 [Oryza sativa (japonica cultivar-gro... 182 6e-45
ref|NP_173353.1| hypothetical protein; protein id: At1g19190.1 [... 181 2e-44
ref|NP_197112.1| putative protein; protein id: At5g16080.1, supp... 179 5e-44
>gb|AAN77692.1|AF487826_1 putative serine hydrolase [Vitis vinifera]
Length = 310
Score = 374 bits (960), Expect = e-102
Identities = 187/309 (60%), Positives = 236/309 (75%), Gaps = 1/309 (0%)
Frame = -2
Query: 1088 EAPDFLQVYSDGLVKRFDPETVPASS-KPSNGYKSKDVVIDPSKPITGRIFLPHFENQQN 912
EA + +V SDG +KR + E+ PAS+ SNGYKSKDV+I+ +KPI+ RIFLP
Sbjct: 6 EASAYFKVLSDGSIKRVEWESAPASNDSSSNGYKSKDVIINSTKPISARIFLPDVPGSSG 65
Query: 911 LLPVLVYFHGGGFCIGSTTWLGYHHFLGDFSVASQSIVLSVDYRLAPEHRLPAAYEDCYV 732
LPVLVYFHGGGFC+GSTTW GYH FLGDF+VASQSIVLSVDYR APE+RLP AY+DCY
Sbjct: 66 RLPVLVYFHGGGFCLGSTTWFGYHTFLGDFAVASQSIVLSVDYRHAPENRLPIAYDDCYS 125
Query: 731 ALQWLAEQASSEPWLQGADISRVFLSGESAGGNIAHHVFVKTTQRNNDVLVLPI*GLMVI 552
+L+WL+ Q SSEPWL+ AD+SRVFLSG+SAGGNI H+V ++T Q + V I GL++I
Sbjct: 126 SLEWLSCQVSSEPWLERADLSRVFLSGDSAGGNIVHNVALRTIQEQSCDQV-KIKGLLLI 184
Query: 551 HPYFGSEERTEKEEAVADDDKECVAMNDMFWRLSIPEDSNRDYFGCNFEKHDVDASVWSQ 372
HP+FGSEER EKE A + E +A+ D W+LS+PE SNRD++ CN+E ++ + W +
Sbjct: 185 HPFFGSEERIEKER--AGGEAENLALTDWMWKLSLPEGSNRDHYWCNYEMAELSRAEWCR 242
Query: 371 FPGIEVYVAGRDFLKERGVMYAEFLKKKGVKEVKLVEAKEEPHIFHIFYPQSDATRLLQS 192
FP VYVAG DFLKERGVMYA FL+K GV EVKLVEA+ E H++H+ +P+S+ATRLLQ
Sbjct: 243 FPPAVVYVAGLDFLKERGVMYAAFLEKNGV-EVKLVEAEGEKHVYHMLHPESEATRLLQK 301
Query: 191 QMSHFMKRY 165
QMS F+ +
Sbjct: 302 QMSEFIHNF 310
>gb|AAD04946.2| PrMC3 [Pinus radiata]
Length = 319
Score = 303 bits (777), Expect = 2e-81
Identities = 162/311 (52%), Positives = 210/311 (67%), Gaps = 3/311 (0%)
Frame = -2
Query: 1088 EAPDFLQVYSDGLVKRFDPETVPASSK-PSNGYKSKDVVIDPSKPITGRIFLPHFENQQN 912
+ P F+QVY DG V RFD PAS + S+G +SKDVVIDP K I+ R+FLP
Sbjct: 10 DVPGFIQVYEDGFVARFDHRLTPASPQVASDGARSKDVVIDPVKGISARLFLPAELPLAQ 69
Query: 911 LLPVLVYFHGGGFCIGSTTWLGYHHFLGDFSVASQSIVLSVDYRLAPEHRLPAAYEDCYV 732
LP+L YFHGGGFCIG+T W GYH FL + ++++V+SVDYRLAPEHRLPAAY+DC+
Sbjct: 70 KLPLLFYFHGGGFCIGTTAWEGYHLFLSLLAATTRALVISVDYRLAPEHRLPAAYDDCFD 129
Query: 731 ALQWLAE-QASSEPWLQG-ADISRVFLSGESAGGNIAHHVFVKTTQRNNDVLVLPI*GLM 558
A++W+A +EPWL AD R FL+GESAGGNIAH V +T + D+ L I GL+
Sbjct: 130 AVEWVASGGGKAEPWLDAHADYGRCFLAGESAGGNIAHVVGSRTA--DQDLGPLKIRGLI 187
Query: 557 VIHPYFGSEERTEKEEAVADDDKECVAMNDMFWRLSIPEDSNRDYFGCNFEKHDVDASVW 378
VIHPYFGSEER E E+ A DD + +ND+FWRL++P S+RDY CN
Sbjct: 188 VIHPYFGSEERIECEKVAAGDDAAALELNDLFWRLALPPGSDRDYPTCNPRGPRSADLRK 247
Query: 377 SQFPGIEVYVAGRDFLKERGVMYAEFLKKKGVKEVKLVEAKEEPHIFHIFYPQSDATRLL 198
P + V VAG D LK RG++Y E L+ G KE +L+EA+ E H +H+F+P+S+ATRLL
Sbjct: 248 VPLPPVLVTVAGLDLLKTRGLLYYELLQSCG-KEAELMEAEGEIHAYHVFHPRSEATRLL 306
Query: 197 QSQMSHFMKRY 165
Q +MS F+ R+
Sbjct: 307 QERMSQFIHRF 317
>dbj|BAB44070.1| P0030H07.40 [Oryza sativa (japonica cultivar-group)]
gi|15528618|dbj|BAB64639.1| P0011G08.10 [Oryza sativa
(japonica cultivar-group)]
Length = 337
Score = 182 bits (463), Expect = 6e-45
Identities = 119/313 (38%), Positives = 163/313 (52%), Gaps = 11/313 (3%)
Frame = -2
Query: 1073 LQVYSDGLVKRFDPETVPASSKPSN------GYKSKDVVIDPSKPITGRIFLPHFENQQN 912
+Q+ SDG V R D + + P G + KD+V D + + R++ P
Sbjct: 19 IQLLSDGTVVRSDAGSGAGALLPPEDFPDVPGVQWKDLVYDATHGLKLRVYRPPTAGDAE 78
Query: 911 LLPVLVYFHGGGFCIGSTTWLGYHHFLGDFSVASQSIVLSVDYRLAPEHRLPAAYEDCYV 732
LPVLV FHGGG+C+G+ +H + +++VLS DYRL PEHRLPAA +D
Sbjct: 79 RLPVLVCFHGGGYCLGTFEKPSFHCCCQRLASELRAVVLSADYRLGPEHRLPAAIDDGAA 138
Query: 731 ALQWLAEQASSEP----WL-QGADISRVFLSGESAGGNIAHHVFVKTTQRNNDVLVLPI* 567
L WL +QA S P WL + AD +RVF++GESAGGN++HHV V V L +
Sbjct: 139 VLSWLRDQAMSGPGADSWLAESADFARVFVAGESAGGNMSHHVAVLIGSGQLTVDPLRVA 198
Query: 566 GLMVIHPYFGSEERTEKEEAVADDDKECVAMNDMFWRLSIPEDSNRDYFGCNFEKHDVDA 387
G M++ P+FG ER E M+D WRLS+PE + RD+ N D +
Sbjct: 199 GYMLLTPFFGGVERAPSEAEPPAGAFFTPDMSDKLWRLSLPEGATRDHPVANPFGPDSPS 258
Query: 386 SVWSQFPGIEVYVAGRDFLKERGVMYAEFLKKKGVKEVKLVEAKEEPHIFHIFYPQSDAT 207
FP + V VAGRD L +R V YA LK+ K V+LV +EE H+F P S+
Sbjct: 259 LAAVAFPPVLVVVAGRDILHDRTVHYAARLKEM-EKPVELVTFEEEKHLFLSLQPWSEPA 317
Query: 206 RLLQSQMSHFMKR 168
L M F+ +
Sbjct: 318 NELIRVMKRFIHK 330
>ref|NP_173353.1| hypothetical protein; protein id: At1g19190.1 [Arabidopsis thaliana]
gi|25518494|pir||D86325 hypothetical protein T29M8.6 -
Arabidopsis thaliana gi|8954057|gb|AAF82230.1|AC069143_6
Contains similarity to a PrMC3 from Pinus radiata
gb|AF110333. [Arabidopsis thaliana]
Length = 318
Score = 181 bits (459), Expect = 2e-44
Identities = 114/314 (36%), Positives = 170/314 (53%), Gaps = 14/314 (4%)
Frame = -2
Query: 1070 QVYSDGLVKRFDPET-VPASSKPSNGYKSKDVVIDPSKPITGRIFLPH---FENQQNLLP 903
+++ +G ++R PET VP S P NG SKD V P K ++ RI+LP +E + +P
Sbjct: 14 RIFKNGGIERLVPETFVPPSLNPENGVVSKDAVYSPEKNLSLRIYLPQNSVYETGEKKIP 73
Query: 902 VLVYFHGGGFCIGSTTWLGYHHFLGDFSVASQSIVLSVDYRLAPEHRLPAAYEDCYVALQ 723
+LVYFHGGGF + + YH FL A+ I +SV+YR APEH +P YED + A+Q
Sbjct: 74 LLVYFHGGGFIMETAFSPIYHTFLTSAVSATDCIAVSVEYRRAPEHPIPTLYEDSWDAIQ 133
Query: 722 WL---AEQASSEPWL-QGADISRVFLSGESAGGNIAHHVFVKTTQRNNDVLVLPI*GLMV 555
W+ ++ E WL + AD S+VFL+G+SAG NIAHH+ ++ + I G+++
Sbjct: 134 WIFTHITRSGPEDWLNKHADFSKVFLAGDSAGANIAHHMAIRVDKEKLPPENFKISGMIL 193
Query: 554 IHPYFGSEERTEKEEAVADDDKECVAMNDMFWRLSIPEDSNRDYFGCNFEKHDVDASVWS 375
HPYF S+ E+ E E + + WR++ P+ N E ++ V S
Sbjct: 194 FHPYFLSKALIEEMEV------EAMRYYERLWRIASPDSGN------GVEDPWINV-VGS 240
Query: 374 QFPG-----IEVYVAGRDFLKERGVMYAEFLKKKG-VKEVKLVEAKEEPHIFHIFYPQSD 213
G + V VAG D L G Y L+K G + +VK++E KEE H+FH+ P S+
Sbjct: 241 DLTGLGCRRVLVMVAGNDVLARGGWSYVAELEKSGWIGKVKVMETKEEGHVFHLRDPDSE 300
Query: 212 ATRLLQSQMSHFMK 171
R + + F+K
Sbjct: 301 NARRVLRNFAEFLK 314
>ref|NP_197112.1| putative protein; protein id: At5g16080.1, supported by cDNA: 37170.,
supported by cDNA: gi_13878128, supported by cDNA:
gi_21280966 [Arabidopsis thaliana]
gi|11357575|pir||T51391 hypothetical protein F1N13_220 -
Arabidopsis thaliana gi|9755654|emb|CAC01807.1| putative
protein [Arabidopsis thaliana]
gi|13878129|gb|AAK44142.1|AF370327_1 unknown protein
[Arabidopsis thaliana] gi|21280967|gb|AAM44955.1| unknown
protein [Arabidopsis thaliana]
Length = 344
Score = 179 bits (455), Expect = 5e-44
Identities = 108/298 (36%), Positives = 167/298 (55%), Gaps = 12/298 (4%)
Frame = -2
Query: 1088 EAPDFLQVYSDGLVKRFDPETVPASSK---PSNGYKSKDVVIDPSKPITGRIFLPHFE-- 924
E ++V++DG V+R P VP S PS+ + D+ + S R+++P
Sbjct: 31 EIEGLIKVFNDGCVER--PPIVPIVSPTIHPSSKATAFDIKL--SNDTWTRVYIPDAAAA 86
Query: 923 NQQNLLPVLVYFHGGGFCIGSTTWLGYHHFLGDFSVASQSIVLSVDYRLAPEHRLPAAYE 744
+ LP+LVYFHGGGFC+GS W YH FL +V ++ +++SV+YRLAPEHRLPAAY+
Sbjct: 87 SPSVTLPLLVYFHGGGFCVGSAAWSCYHDFLTSLAVKARCVIVSVNYRLAPEHRLPAAYD 146
Query: 743 DCYVALQWLAEQASS-----EPWLQGADISRVFLSGESAGGNIAHHVFVKTTQRNNDVLV 579
D + WL +Q S WL ++S VFL+G+SAG NIA+ V V+
Sbjct: 147 DGVNVVSWLVKQQISTGGGYPSWLSKCNLSNVFLAGDSAGANIAYQVAVRIMASGKYANT 206
Query: 578 LPI*GLMVIHPYFGSEERT--EKEEAVADDDKECVAMNDMFWRLSIPEDSNRDYFGCNFE 405
L + G+++IHP+FG E RT EK++ ++ +D +WRL++P ++RD+ CN
Sbjct: 207 LHLKGIILIHPFFGGESRTSSEKQQHHTKSSALTLSASDAYWRLALPRGASRDHPWCN-- 264
Query: 404 KHDVDASVWSQFPGIEVYVAGRDFLKERGVMYAEFLKKKGVKEVKLVEAKEEPHIFHI 231
+ +S ++ P V++A D LKER + + ++ G K V+ + H FHI
Sbjct: 265 --PLMSSAGAKLPTTMVFMAEFDILKERNLEMCKVMRSHG-KRVEGIVHGGVGHAFHI 319
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,018,356,855
Number of Sequences: 1393205
Number of extensions: 24806835
Number of successful extensions: 77173
Number of sequences better than 10.0: 1017
Number of HSP's better than 10.0 without gapping: 69994
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76211
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 65340192036
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)