Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC019090A_C01 KMC019090A_c01
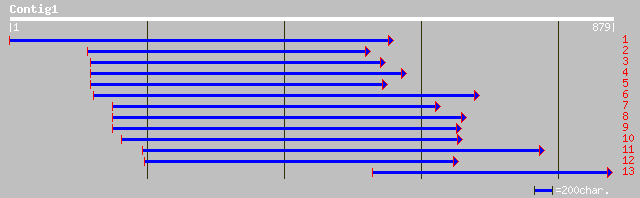
(874 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T15054 anther-specific protein - wood tobacco gi|2326772|em... 309 3e-83
emb|CAA50308.1| anther specific protein [Brassica napus] 303 2e-81
pir||S32145 anther-specific protein - rape (fragment) 301 8e-81
ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabi... 299 3e-80
pir||S26252 anther-specific protein (clone BA42) - rape (fragmen... 290 1e-77
>pir||T15054 anther-specific protein - wood tobacco gi|2326772|emb|CAA74847.1|
anther-specific protein [Nicotiana sylvestris]
gi|2326774|emb|CAA74846.1| anther-specific protein
[Nicotiana sylvestris]
Length = 363
Score = 309 bits (792), Expect = 3e-83
Identities = 150/191 (78%), Positives = 172/191 (89%), Gaps = 4/191 (2%)
Frame = -3
Query: 872 ILGFRPPNKARPYDLVGAALFGDGAAAAIIGANPVSGEESPFLELNYAIQKFLPGTQEVI 693
ILGFRPPNKARPYDLVGAALFGDGAAA IIG P+ G+ESPF+ELN+A Q+FLPGT VI
Sbjct: 173 ILGFRPPNKARPYDLVGAALFGDGAAAVIIGTEPIMGKESPFMELNFATQQFLPGTNNVI 232
Query: 692 DGRLSEEGINFKLGRDLPQKIEDNIEGFCRKLMAKSNVED--FNDLFWAVHPGGPAILNR 519
DGRL+EEGINFKLGRDLP+KI+DNIE FC+K++AK+++ + +NDLFWAVHPGGPAILNR
Sbjct: 233 DGRLTEEGINFKLGRDLPEKIQDNIEEFCKKIIAKADLREAKYNDLFWAVHPGGPAILNR 292
Query: 518 LETKLKLSEEKLECSRKALINYGNVSSNTIFYVMEYMREFL--KEDGGEEWGLGLAFGPG 345
LE LKL EKL+CSR+AL++YGNVSSNTIFYVMEYMRE L K++GGEEWGL LAFGPG
Sbjct: 293 LENTLKLQSEKLDCSRRALMDYGNVSSNTIFYVMEYMREELKNKKNGGEEWGLALAFGPG 352
Query: 344 ITFEGILLRSL 312
ITFEGILLRSL
Sbjct: 353 ITFEGILLRSL 363
>emb|CAA50308.1| anther specific protein [Brassica napus]
Length = 191
Score = 303 bits (777), Expect = 2e-81
Identities = 149/189 (78%), Positives = 170/189 (89%), Gaps = 4/189 (2%)
Frame = -3
Query: 866 GFRPPNKARPYDLVGAALFGDGAAAAIIGANPVSGEESPFLELNYAIQKFLPGTQEVIDG 687
GFRPPNKARPYDLVGAALFGDGAAA IIGA+P+ E SPF+EL+YA+Q+F+PGTQ VIDG
Sbjct: 1 GFRPPNKARPYDLVGAALFGDGAAAVIIGADPLPSE-SPFMELHYAVQQFVPGTQTVIDG 59
Query: 686 RLSEEGINFKLGRDLPQKIEDNIEGFCRKLMAKSNVE--DFNDLFWAVHPGGPAILNRLE 513
RL+EEGINFKLGRDLPQKIE+NIE FC+KLM K+ + +FND+FW VHPGGPAILNRLE
Sbjct: 60 RLTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDDSMEFNDMFWTVHPGGPAILNRLE 119
Query: 512 TKLKLSEEKLECSRKALINYGNVSSNTIFYVMEYMREFLKE--DGGEEWGLGLAFGPGIT 339
TKLKL EKLECSR+AL++YGNVSSNTI YVMEYMR+ LK+ DG +EWGLGLAFGPGIT
Sbjct: 120 TKLKLGREKLECSRRALVDYGNVSSNTILYVMEYMRDELKKKGDGAQEWGLGLAFGPGIT 179
Query: 338 FEGILLRSL 312
FEG+LLRSL
Sbjct: 180 FEGLLLRSL 188
>pir||S32145 anther-specific protein - rape (fragment)
Length = 190
Score = 301 bits (771), Expect = 8e-81
Identities = 148/188 (78%), Positives = 169/188 (89%), Gaps = 4/188 (2%)
Frame = -3
Query: 863 FRPPNKARPYDLVGAALFGDGAAAAIIGANPVSGEESPFLELNYAIQKFLPGTQEVIDGR 684
FRPPNKARPYDLVGAALFGDGAAA IIGA+P+ E SPF+EL+YA+Q+F+PGTQ VIDGR
Sbjct: 1 FRPPNKARPYDLVGAALFGDGAAAVIIGADPLPSE-SPFMELHYAVQQFVPGTQTVIDGR 59
Query: 683 LSEEGINFKLGRDLPQKIEDNIEGFCRKLMAKSNVE--DFNDLFWAVHPGGPAILNRLET 510
L+EEGINFKLGRDLPQKIE+NIE FC+KLM K+ + +FND+FW VHPGGPAILNRLET
Sbjct: 60 LTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDDSMEFNDMFWTVHPGGPAILNRLET 119
Query: 509 KLKLSEEKLECSRKALINYGNVSSNTIFYVMEYMREFLKE--DGGEEWGLGLAFGPGITF 336
KLKL EKLECSR+AL++YGNVSSNTI YVMEYMR+ LK+ DG +EWGLGLAFGPGITF
Sbjct: 120 KLKLGREKLECSRRALVDYGNVSSNTILYVMEYMRDELKKKGDGAQEWGLGLAFGPGITF 179
Query: 335 EGILLRSL 312
EG+LLRSL
Sbjct: 180 EGLLLRSL 187
>ref|NP_171707.1| unknown protein; protein id: At1g02050.1 [Arabidopsis thaliana]
gi|25286664|pir||E86152 chalcone synthase homolog
T7I23.4 - Arabidopsis thaliana gi|2317904|gb|AAC24368.1|
Similar to rice chalcone synthase homolog,
gp|U90341|2507617 and anther specific protein,
gp|Y14507|2326772 [Arabidopsis thaliana]
Length = 395
Score = 299 bits (766), Expect = 3e-80
Identities = 149/191 (78%), Positives = 171/191 (89%), Gaps = 4/191 (2%)
Frame = -3
Query: 872 ILGFRPPNKARPYDLVGAALFGDGAAAAIIGANPVSGEESPFLELNYAIQKFLPGTQEVI 693
ILGFRPPNKARPYDLVGAALFGDGAAA IIGA+P E +PF+EL+YA+Q+FLPGTQ VI
Sbjct: 203 ILGFRPPNKARPYDLVGAALFGDGAAAVIIGADPRECE-APFMELHYAVQQFLPGTQNVI 261
Query: 692 DGRLSEEGINFKLGRDLPQKIEDNIEGFCRKLMAKSNVE--DFNDLFWAVHPGGPAILNR 519
+GRL+EEGINFKLGRDLPQKIE+NIE FC+KLM K+ E +FND+FWAVHPGGPAILNR
Sbjct: 262 EGRLTEEGINFKLGRDLPQKIEENIEEFCKKLMGKAGDESMEFNDMFWAVHPGGPAILNR 321
Query: 518 LETKLKLSEEKLECSRKALINYGNVSSNTIFYVMEYMREFLKE--DGGEEWGLGLAFGPG 345
LETKLKL +EKLE SR+AL++YGNVSSNTI YVMEYMR+ LK+ D +EWGLGLAFGPG
Sbjct: 322 LETKLKLEKEKLESSRRALVDYGNVSSNTILYVMEYMRDELKKKGDAAQEWGLGLAFGPG 381
Query: 344 ITFEGILLRSL 312
ITFEG+L+RSL
Sbjct: 382 ITFEGLLIRSL 392
>pir||S26252 anther-specific protein (clone BA42) - rape (fragment)
gi|258236|gb|AAB23817.1| chalcone synthase homolog
{C-terminal} [Brassica napus, cv. Westar, Peptide
Partial, 222 aa]
Length = 222
Score = 290 bits (743), Expect = 1e-77
Identities = 146/191 (76%), Positives = 167/191 (86%), Gaps = 4/191 (2%)
Frame = -3
Query: 872 ILGFRPPNKARPYDLVGAALFGDGAAAAIIGANPVSGEESPFLELNYAIQKFLPGTQEVI 693
+LGFRPPNKARPYDLVGAALFGDGAAA IIGA+P E SPF+EL+YA+Q+FLPGTQ VI
Sbjct: 33 VLGFRPPNKARPYDLVGAALFGDGAAALIIGADPTESE-SPFMELHYALQQFLPGTQAVI 91
Query: 692 DGRLSEEGINFKLGRDLPQKIEDNIEGFCRKLMAK--SNVEDFNDLFWAVHPGGPAILNR 519
DGRLSEEGI+FKLGR+LPQKIEDNIE FC+KL+AK S + NDLFWAVHP GPAILN
Sbjct: 92 DGRLSEEGISFKLGRELPQKIEDNIEEFCKKLVAKAGSGSLELNDLFWAVHPCGPAILNG 151
Query: 518 LETKLKLSEEKLECSRKALINYGNVSSNTIFYVMEYMREFLKEDG--GEEWGLGLAFGPG 345
LETKLKL EKLECSR+AL++YGN SSNTIFY+M+ +R+ L++ G GEEWGLGLAFGPG
Sbjct: 152 LETKLKLKPEKLECSRQALVDYGNASSNTIFYIMDKVRDELEKKGRSGEEWGLGLAFGPG 211
Query: 344 ITFEGILLRSL 312
ITFEG L+RSL
Sbjct: 212 ITFEGFLMRSL 222
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 803,472,664
Number of Sequences: 1393205
Number of extensions: 19090177
Number of successful extensions: 82789
Number of sequences better than 10.0: 627
Number of HSP's better than 10.0 without gapping: 65456
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 79497
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 46824663816
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)