Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
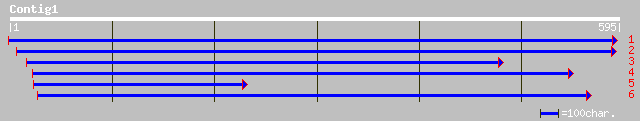
Query= KMC019088A_C01 KMC019088A_c01
(594 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_192230.1| xyloglucan endotransglycosylase, putative; prot... 246 2e-64
gb|AAM62971.1| putative xyloglucan endotransglycosylase [Arabido... 246 2e-64
ref|NP_196891.1| xyloglucan endotransglycosylase (EXGT-A4); prot... 184 7e-46
dbj|BAB17788.1| xyloglucan endotransglycosylase [Pisum sativum] 184 1e-45
dbj|BAA34946.1| EXGT1 [Pisum sativum] 184 1e-45
>ref|NP_192230.1| xyloglucan endotransglycosylase, putative; protein id: At4g03210.1,
supported by cDNA: 17748., supported by cDNA:
gi_14994278, supported by cDNA: gi_18252836 [Arabidopsis
thaliana] gi|25313540|pir||G85040 probable xyloglucan
endotransglycosylase [imported] - Arabidopsis thaliana
gi|4262149|gb|AAD14449.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|7270191|emb|CAB77806.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|14994279|gb|AAK73274.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|18252837|gb|AAL62345.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
gi|25084277|gb|AAN72210.1| putative xyloglucan
endotransglycosylase [Arabidopsis thaliana]
Length = 290
Score = 246 bits (628), Expect = 2e-64
Identities = 112/141 (79%), Positives = 129/141 (91%)
Frame = +1
Query: 172 VSSAKFDELFQSSWALDHFIHEGEQIKLQLDKYSGAGFESKKKYMFGRVSLNLKLVEGDS 351
VS AKFDEL++SSWA+DH ++EGE KL+LD YSGAGFES+ KY+FG+VS+ +KLVEGDS
Sbjct: 24 VSGAKFDELYRSSWAMDHCVNEGEVTKLKLDNYSGAGFESRSKYLFGKVSIQIKLVEGDS 83
Query: 352 AGTVTAFYMSSEGANHNEFDFEFLGNTTGEPYSVQTNVYVNGVGNREQRLNLWFDPTKEF 531
AGTVTAFYMSS+G NHNEFDFEFLGNTTGEPY VQTN+YVNGVGNREQRLNLWFDPT EF
Sbjct: 84 AGTVTAFYMSSDGPNHNEFDFEFLGNTTGEPYIVQTNIYVNGVGNREQRLNLWFDPTTEF 143
Query: 532 HSYSIFWNQRQVVFLVDETPI 594
H+YSI W++R VVF+VDETPI
Sbjct: 144 HTYSILWSKRSVVFMVDETPI 164
>gb|AAM62971.1| putative xyloglucan endotransglycosylase [Arabidopsis thaliana]
Length = 287
Score = 246 bits (628), Expect = 2e-64
Identities = 112/141 (79%), Positives = 129/141 (91%)
Frame = +1
Query: 172 VSSAKFDELFQSSWALDHFIHEGEQIKLQLDKYSGAGFESKKKYMFGRVSLNLKLVEGDS 351
VS AKFDEL++SSWA+DH ++EGE KL+LD YSGAGFES+ KY+FG+VS+ +KLVEGDS
Sbjct: 21 VSGAKFDELYRSSWAMDHCVNEGEVTKLKLDNYSGAGFESRSKYLFGKVSIQIKLVEGDS 80
Query: 352 AGTVTAFYMSSEGANHNEFDFEFLGNTTGEPYSVQTNVYVNGVGNREQRLNLWFDPTKEF 531
AGTVTAFYMSS+G NHNEFDFEFLGNTTGEPY VQTN+YVNGVGNREQRLNLWFDPT EF
Sbjct: 81 AGTVTAFYMSSDGPNHNEFDFEFLGNTTGEPYIVQTNIYVNGVGNREQRLNLWFDPTTEF 140
Query: 532 HSYSIFWNQRQVVFLVDETPI 594
H+YSI W++R VVF+VDETPI
Sbjct: 141 HTYSILWSKRSVVFMVDETPI 161
>ref|NP_196891.1| xyloglucan endotransglycosylase (EXGT-A4); protein id: At5g13870.1
[Arabidopsis thaliana] gi|5139002|dbj|BAA81669.1|
endoxyloglucan transferase [Arabidopsis thaliana]
gi|5533315|gb|AAD45126.1|AF163822_1 endoxyloglucan
transferase [Arabidopsis thaliana]
gi|10177653|dbj|BAB11115.1| endoxyloglucan transferase
[Arabidopsis thaliana]
Length = 293
Score = 184 bits (467), Expect = 7e-46
Identities = 79/138 (57%), Positives = 107/138 (77%), Gaps = 2/138 (1%)
Frame = +1
Query: 187 FDELFQSSWALDH--FIHEGEQIKLQLDKYSGAGFESKKKYMFGRVSLNLKLVEGDSAGT 360
F + +WA DH +++ G ++ L LDKY+G GF+SK Y+FG S+++K+V GDSAGT
Sbjct: 32 FGRNYFPTWAFDHIKYLNGGSEVHLVLDKYTGTGFQSKGSYLFGHFSMHIKMVAGDSAGT 91
Query: 361 VTAFYMSSEGANHNEFDFEFLGNTTGEPYSVQTNVYVNGVGNREQRLNLWFDPTKEFHSY 540
VTAFY+SS+ + H+E DFEFLGN TG+PY +QTNV+ G GNREQR+NLWFDP+K++HSY
Sbjct: 92 VTAFYLSSQNSEHDEIDFEFLGNRTGQPYILQTNVFTGGAGNREQRINLWFDPSKDYHSY 151
Query: 541 SIFWNQRQVVFLVDETPI 594
S+ WN Q+VF VD+ PI
Sbjct: 152 SVLWNMYQIVFFVDDVPI 169
>dbj|BAB17788.1| xyloglucan endotransglycosylase [Pisum sativum]
Length = 293
Score = 184 bits (466), Expect = 1e-45
Identities = 88/162 (54%), Positives = 113/162 (69%), Gaps = 13/162 (8%)
Frame = +1
Query: 148 VGLVMVGLVSSA-----------KFDELFQSSWALDH--FIHEGEQIKLQLDKYSGAGFE 288
V L++ LVSS+ F + +WA DH + + G +I+L LDK +G GF+
Sbjct: 8 VCLILASLVSSSFCALPRKPVDVPFGRNYYPTWAFDHIKYFNGGSEIQLHLDKSTGTGFQ 67
Query: 289 SKKKYMFGRVSLNLKLVEGDSAGTVTAFYMSSEGANHNEFDFEFLGNTTGEPYSVQTNVY 468
SK Y+FG S+N+K+V GDSAGTVTAFY+SS+ A H+E DFEFLGN TG+PY +QTNV+
Sbjct: 68 SKGSYLFGHFSMNIKMVPGDSAGTVTAFYLSSQNAEHDEIDFEFLGNRTGQPYILQTNVF 127
Query: 469 VNGVGNREQRLNLWFDPTKEFHSYSIFWNQRQVVFLVDETPI 594
G GN+EQR+ LWFDPTKEFH YSI WN Q+VF VD+ PI
Sbjct: 128 TGGQGNKEQRIFLWFDPTKEFHRYSILWNMYQIVFYVDDVPI 169
>dbj|BAA34946.1| EXGT1 [Pisum sativum]
Length = 293
Score = 184 bits (466), Expect = 1e-45
Identities = 88/162 (54%), Positives = 113/162 (69%), Gaps = 13/162 (8%)
Frame = +1
Query: 148 VGLVMVGLVSSA-----------KFDELFQSSWALDH--FIHEGEQIKLQLDKYSGAGFE 288
V L++ LVSS+ F + +WA DH + + G +I+L LDK +G GF+
Sbjct: 8 VCLILASLVSSSFCALPRKPVDVPFGRNYYPTWAFDHIKYFNGGSEIQLHLDKSTGTGFQ 67
Query: 289 SKKKYMFGRVSLNLKLVEGDSAGTVTAFYMSSEGANHNEFDFEFLGNTTGEPYSVQTNVY 468
SK Y+FG S+N+K+V GDSAGTVTAFY+SS+ A H+E DFEFLGN TG+PY +QTNV+
Sbjct: 68 SKGSYLFGHFSMNIKMVPGDSAGTVTAFYLSSQNAEHDEIDFEFLGNRTGQPYILQTNVF 127
Query: 469 VNGVGNREQRLNLWFDPTKEFHSYSIFWNQRQVVFLVDETPI 594
G GN+EQR+ LWFDPTKEFH YSI WN Q+VF VD+ PI
Sbjct: 128 TGGQGNKEQRIFLWFDPTKEFHRYSILWNMYQIVFYVDDVPI 169
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 537,397,298
Number of Sequences: 1393205
Number of extensions: 12257376
Number of successful extensions: 44096
Number of sequences better than 10.0: 224
Number of HSP's better than 10.0 without gapping: 41884
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43962
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)