Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
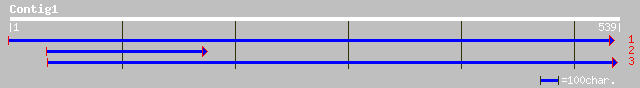
Query= KMC019087A_C01 KMC019087A_c01
(539 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_193739.1| hypothetical protein; protein id: At4g20060.1 [... 55 7e-07
gb|EAA18721.1| glutamine-asparagine rich protein [Plasmodium yoe... 54 9e-07
ref|NP_473081.2| hypothetical protein [Plasmodium falciparum 3D7... 49 3e-05
pir||F71606 probable multiple transmembrane domain protein PFB07... 49 3e-05
gb|AAA17387.1| glutamine-asparagine rich protein 49 4e-05
>ref|NP_193739.1| hypothetical protein; protein id: At4g20060.1 [Arabidopsis
thaliana] gi|7452440|pir||T04890 hypothetical protein
F18F4.160 - Arabidopsis thaliana
gi|2827660|emb|CAA16614.1| hypothetical protein
[Arabidopsis thaliana] gi|7268801|emb|CAB79006.1|
hypothetical protein [Arabidopsis thaliana]
Length = 1134
Score = 54.7 bits (130), Expect = 7e-07
Identities = 28/50 (56%), Positives = 33/50 (66%)
Frame = +3
Query: 372 MEPTCAACAMEWSIQLEKGLRSSKPGVPVKTILKLGPHLQLWSREFESGI 521
ME AACAMEWSI+LEK LRS V+ IL+ G L+ WS+E ES I
Sbjct: 1 MEKVSAACAMEWSIKLEKSLRSKNSVKAVEAILETGGKLEQWSKEPESAI 50
>gb|EAA18721.1| glutamine-asparagine rich protein [Plasmodium yoelii yoelii]
Length = 978
Score = 54.3 bits (129), Expect = 9e-07
Identities = 36/94 (38%), Positives = 53/94 (56%)
Frame = -3
Query: 294 RLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVE 115
+LKQ +LKQ + + RL Q +LKQ + + RL Q +LKQ + + RL Q +LK EQ +E
Sbjct: 398 KLKQEKLKQEQLERERLAQEKLKQEQLEQERLAQEKLKQEQLERERLAQEKLKQEQ--LE 455
Query: 114 EDSIAQRELHDPEEADLIRESIFAQRELNERASI 13
++ +AQ +L A E +RE ER I
Sbjct: 456 QERLAQEKLKQERLAQEKLEREKLEREKLEREKI 489
Score = 50.1 bits (118), Expect = 2e-05
Identities = 33/93 (35%), Positives = 53/93 (56%), Gaps = 2/93 (2%)
Frame = -3
Query: 294 RLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVE 115
+LKQ +L Q + +LKQ +LKQ + + RL Q +LKQ + + RL Q +LK EQ +E
Sbjct: 383 KLKQEQLAQEKLKQEKLKQEKLKQEQLERERLAQEKLKQEQLEQERLAQEKLKQEQ--LE 440
Query: 114 EDSIAQRELHDP--EEADLIRESIFAQRELNER 22
+ +AQ +L E+ L +E + +R E+
Sbjct: 441 RERLAQEKLKQEQLEQERLAQEKLKQERLAQEK 473
Score = 50.1 bits (118), Expect = 2e-05
Identities = 36/97 (37%), Positives = 51/97 (52%)
Frame = -3
Query: 303 ESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQS 124
E RL Q +LKQ + + RL Q +LKQ + + RL Q +LKQ + + RL Q +LK E+
Sbjct: 410 ERERLAQEKLKQEQLEQERLAQEKLKQEQLERERLAQEKLKQEQLEQERLAQEKLKQERL 469
Query: 123 FVEEDSIAQRELHDPEEADLIRESIFAQRELNERASI 13
E+ + E E + RE I +RE ER I
Sbjct: 470 AQEKLEREKLEREKLEREKIEREKI--EREKIEREKI 504
Score = 43.9 bits (102), Expect = 0.001
Identities = 31/90 (34%), Positives = 54/90 (59%)
Frame = -3
Query: 294 RLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVE 115
+L++ +L Q + +LKQ +L Q + +LKQ +LKQ + + RL Q +LK EQ +E
Sbjct: 368 QLEREKLAQEKLKQEKLKQEQLAQEKLKQEKLKQEKLKQEQLERERLAQEKLKQEQ--LE 425
Query: 114 EDSIAQRELHDPEEADLIRESIFAQRELNE 25
++ +AQ +L ++ L RE + AQ +L +
Sbjct: 426 QERLAQEKL---KQEQLERERL-AQEKLKQ 451
Score = 43.1 bits (100), Expect = 0.002
Identities = 32/99 (32%), Positives = 54/99 (54%)
Frame = -3
Query: 303 ESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQS 124
E RL Q +LKQ + + RL Q +LKQ + + RL Q +LKQ +L++ +L+ E+
Sbjct: 425 EQERLAQEKLKQEQLERERLAQEKLKQEQLEQERLAQEKLKQERLAQEKLEREKLEREK- 483
Query: 123 FVEEDSIAQRELHDPEEADLIRESIFAQRELNERASIGE 7
+E + I + ++ E + RE I +RE E+ + E
Sbjct: 484 -LEREKIEREKI---EREKIEREKI--EREQLEKKKLEE 516
Score = 42.4 bits (98), Expect = 0.004
Identities = 32/97 (32%), Positives = 54/97 (54%), Gaps = 2/97 (2%)
Frame = -3
Query: 309 RRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHE 130
R E +LKQ +LKQ + +L Q RL Q + +LKQ +L++ + +LKQ +LK E
Sbjct: 328 RLEEEKLKQEKLKQEKLAQEKLAQERLAQEKLAQEKLKQEQLEREKLAQEKLKQEKLKQE 387
Query: 129 QSFVEEDSIAQRELHDP--EEADLIRESIFAQRELNE 25
Q + ++ + Q +L ++ L RE + AQ +L +
Sbjct: 388 Q--LAQEKLKQEKLKQEKLKQEQLERERL-AQEKLKQ 421
Score = 41.2 bits (95), Expect = 0.008
Identities = 26/103 (25%), Positives = 51/103 (49%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLK 136
++ RE + +++ +Q E+ + K+L K+ EE + K+L K+ EE+ ++ R+K
Sbjct: 493 KIEREKIEREKIEREQLEKKKLEEKRLEEKRLEEKRLEEKRLEEKRIEEEKRIKEEQRIK 552
Query: 135 HEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNERASIGE 7
EQ EE I + + E+ + I ++ + E I E
Sbjct: 553 EEQRIEEEKRIKEEKRIKEEQRIEEEKRIKEEKRIKEEKRIKE 595
Score = 39.3 bits (90), Expect = 0.031
Identities = 27/96 (28%), Positives = 52/96 (54%), Gaps = 2/96 (2%)
Frame = -3
Query: 303 ESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQS 124
E RL++ +LKQ + +L Q +L Q +L Q +LKQ + + +L Q +LK E+
Sbjct: 325 EEKRLEEEKLKQEKLKQEKLAQEKLAQERLAQEKLAQEKLKQEQLEREKLAQEKLKQEK- 383
Query: 123 FVEEDSIAQRELHDP--EEADLIRESIFAQRELNER 22
++++ +AQ +L ++ L +E + +R E+
Sbjct: 384 -LKQEQLAQEKLKQEKLKQEKLKQEQLERERLAQEK 418
Score = 38.9 bits (89), Expect = 0.040
Identities = 27/106 (25%), Positives = 58/106 (54%), Gaps = 5/106 (4%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQRE-EDSVRLKQ---LRLKQREEDSVRLKQ 148
R + E +L++ RL+++ + RL++ RL+++ E+ R+K+ ++ +QR E+ R+K+
Sbjct: 506 REQLEKKKLEEKRLEEKRLEEKRLEEKRLEEKRIEEEKRIKEEQRIKEEQRIEEEKRIKE 565
Query: 147 V-RLKHEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNERASI 13
R+K EQ EE I + + E+ + I ++ + E +
Sbjct: 566 EKRIKEEQRIEEEKRIKEEKRIKEEKRIKEEKRIKEEKRIEEEKKL 611
Score = 38.1 bits (87), Expect = 0.069
Identities = 30/106 (28%), Positives = 59/106 (55%), Gaps = 7/106 (6%)
Frame = -3
Query: 309 RRESVRLKQVRLKQRE-EDSVRLKQ---LRLKQREEDSVRLKQ---LRLKQREEDSVRLK 151
R E RL++ RL+++ E+ R+K+ ++ +QR E+ R+K+ ++ +QR E+ R+K
Sbjct: 523 RLEEKRLEEKRLEEKRIEEEKRIKEEQRIKEEQRIEEEKRIKEEKRIKEEQRIEEEKRIK 582
Query: 150 QVRLKHEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNERASI 13
+ + E+ ++E+ + E EE L +E + E ER +I
Sbjct: 583 EEKRIKEEKRIKEEKRIKEEKRIEEEKKLEKEK-KPELENKERGTI 627
Score = 37.4 bits (85), Expect = 0.12
Identities = 23/95 (24%), Positives = 52/95 (54%), Gaps = 1/95 (1%)
Frame = -3
Query: 303 ESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQS 124
E RL Q +LKQ + + RL Q +LKQ +L++ +L++ + + ++++ +++ E+
Sbjct: 440 ERERLAQEKLKQEQLEQERLAQEKLKQERLAQEKLEREKLEREKLEREKIEREKIEREK- 498
Query: 123 FVEEDSIAQRELHDPE-EADLIRESIFAQRELNER 22
+E + I + +L + E + E ++ L E+
Sbjct: 499 -IEREKIEREQLEKKKLEEKRLEEKRLEEKRLEEK 532
Score = 36.6 bits (83), Expect = 0.20
Identities = 26/88 (29%), Positives = 49/88 (55%)
Frame = -3
Query: 288 KQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVEED 109
+Q +L+++ + +LKQ +LKQ + +L Q RL Q +L Q +LK EQ +E +
Sbjct: 320 EQEKLEEKRLEEEKLKQEKLKQEKLAQEKLAQERLAQE-----KLAQEKLKQEQ--LERE 372
Query: 108 SIAQRELHDPEEADLIRESIFAQRELNE 25
+AQ +L + + +++ AQ +L +
Sbjct: 373 KLAQEKL----KQEKLKQEQLAQEKLKQ 396
Score = 36.6 bits (83), Expect = 0.20
Identities = 22/99 (22%), Positives = 55/99 (55%), Gaps = 1/99 (1%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLK 136
R R +LKQ +L+Q +LKQ RL Q + + +L++ +L++ + + ++++ +++
Sbjct: 441 RERLAQEKLKQEQLEQERLAQEKLKQERLAQEKLEREKLEREKLEREKIEREKIEREKIE 500
Query: 135 HEQSFVEEDSIAQRELHDPE-EADLIRESIFAQRELNER 22
E+ +E + + +++L + E + E ++ L E+
Sbjct: 501 REK--IEREQLEKKKLEEKRLEEKRLEEKRLEEKRLEEK 537
>ref|NP_473081.2| hypothetical protein [Plasmodium falciparum 3D7]
gi|23503421|gb|AAC71942.2| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1040
Score = 49.3 bits (116), Expect = 3e-05
Identities = 24/73 (32%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Frame = +2
Query: 89 SSLCAIESSSTKLCS---CFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTES 259
++ C +S C+ C N TC N T ++ C N +C N T ++ C N +C N T +
Sbjct: 391 NNTCNNHTSDNNTCNNHTCDNHTCDNHTSDNNTCNNHTCNNHTCNNHTCNNHTCNNHTCN 450
Query: 260 SSLCFNRTCFNRT 298
+ C N T N T
Sbjct: 451 NHTCNNHTSDNNT 463
Score = 48.9 bits (115), Expect = 4e-05
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Frame = +2
Query: 98 CAIESSSTKLC---SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSL 268
C +S + C +C + TC N T ++ C N + N T ++ C N +C N T ++
Sbjct: 364 CDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHTCDNHTSDNNT 423
Query: 269 CFNRTCFNRT 298
C N TC N T
Sbjct: 424 CNNHTCNNHT 433
Score = 48.9 bits (115), Expect = 4e-05
Identities = 21/53 (39%), Positives = 29/53 (54%)
Frame = +2
Query: 140 NRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
N TC N T ++ C N +C N T ++ C N +C N T ++ C N TC N T
Sbjct: 421 NNTCNNHTCNNHTCNNHTCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNT 473
Score = 43.1 bits (100), Expect = 0.002
Identities = 19/53 (35%), Positives = 26/53 (48%)
Frame = +2
Query: 140 NRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
N TC N T S C + +C + T + N +C N T ++ C N TC N T
Sbjct: 361 NHTCDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHT 413
Score = 41.2 bits (95), Expect = 0.008
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFN 292
+C N TC N T ++ C N +C N T ++ C N +C N T C N T N
Sbjct: 433 TCNNHTCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNT-----CNNHTLGN 481
Score = 40.8 bits (94), Expect = 0.011
Identities = 19/56 (33%), Positives = 28/56 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
+C N T + T S C + +C N T ++ C N + N T ++ C N TC N T
Sbjct: 363 TCDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHTCDNHT 418
Score = 37.7 bits (86), Expect = 0.090
Identities = 18/56 (32%), Positives = 28/56 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
+C N TC N T ++ C N + N T ++ C N +C N T + +N +N T
Sbjct: 438 TCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNTCNNHTLGNPHFYNPHFYNNT 493
>pir||F71606 probable multiple transmembrane domain protein PFB0770c - malaria
parasite (Plasmodium falciparum)
Length = 1122
Score = 49.3 bits (116), Expect = 3e-05
Identities = 24/73 (32%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Frame = +2
Query: 89 SSLCAIESSSTKLCS---CFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTES 259
++ C +S C+ C N TC N T ++ C N +C N T ++ C N +C N T +
Sbjct: 473 NNTCNNHTSDNNTCNNHTCDNHTCDNHTSDNNTCNNHTCNNHTCNNHTCNNHTCNNHTCN 532
Query: 260 SSLCFNRTCFNRT 298
+ C N T N T
Sbjct: 533 NHTCNNHTSDNNT 545
Score = 48.9 bits (115), Expect = 4e-05
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Frame = +2
Query: 98 CAIESSSTKLC---SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSL 268
C +S + C +C + TC N T ++ C N + N T ++ C N +C N T ++
Sbjct: 446 CDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHTCDNHTSDNNT 505
Query: 269 CFNRTCFNRT 298
C N TC N T
Sbjct: 506 CNNHTCNNHT 515
Score = 48.9 bits (115), Expect = 4e-05
Identities = 21/53 (39%), Positives = 29/53 (54%)
Frame = +2
Query: 140 NRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
N TC N T ++ C N +C N T ++ C N +C N T ++ C N TC N T
Sbjct: 503 NNTCNNHTCNNHTCNNHTCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNT 555
Score = 43.1 bits (100), Expect = 0.002
Identities = 19/53 (35%), Positives = 26/53 (48%)
Frame = +2
Query: 140 NRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
N TC N T S C + +C + T + N +C N T ++ C N TC N T
Sbjct: 443 NHTCDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHT 495
Score = 41.2 bits (95), Expect = 0.008
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFN 292
+C N TC N T ++ C N +C N T ++ C N +C N T C N T N
Sbjct: 515 TCNNHTCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNT-----CNNHTLGN 563
Score = 40.8 bits (94), Expect = 0.011
Identities = 19/56 (33%), Positives = 28/56 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
+C N T + T S C + +C N T ++ C N + N T ++ C N TC N T
Sbjct: 445 TCDNHTSDSHTCDSHTCDSHTCDNHTSDNNTCNNHTSDNNTCNNHTCDNHTCDNHT 500
Score = 37.7 bits (86), Expect = 0.090
Identities = 18/56 (32%), Positives = 28/56 (49%)
Frame = +2
Query: 131 SCFNRTCFNRTESSSLCFNRSCFNRTESSSLCFNRSCFNRTESSSLCFNRTCFNRT 298
+C N TC N T ++ C N + N T ++ C N +C N T + +N +N T
Sbjct: 520 TCNNHTCNNHTCNNHTCNNHTSDNNTCNNHTCDNNTCNNHTLGNPHFYNPHFYNNT 575
>gb|AAA17387.1| glutamine-asparagine rich protein
Length = 720
Score = 48.9 bits (115), Expect = 4e-05
Identities = 22/92 (23%), Positives = 63/92 (67%), Gaps = 3/92 (3%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVR-- 142
++++E ++L+Q++ +Q +++ ++ +QL+ +Q +++ ++ +QL+L+Q +++ ++L+Q++
Sbjct: 353 QLKQEQIKLEQLKQEQLKQEQLKQEQLKKEQLKQEQIKQEQLKLEQIKQEQLKLEQLKQE 412
Query: 141 -LKHEQSFVEEDSIAQRELHDPEEADLIRESI 49
LK EQ E+ + Q + + ++ L +E I
Sbjct: 413 ELKQEQLKQEQLKLKQLKQEELKQEQLKQEQI 444
Score = 48.9 bits (115), Expect = 4e-05
Identities = 26/98 (26%), Positives = 65/98 (65%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLK 136
++++E ++LKQ++ ++ +++ ++ +Q++L+Q +++ +LKQ +LKQ + +LKQ ++K
Sbjct: 418 QLKQEQLKLKQLKQEELKQEQLKQEQIKLEQLKQE--QLKQEQLKQEQLKKEQLKQEQIK 475
Query: 135 HEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNER 22
EQ +E+ Q +L ++ +L +E + Q +L ++
Sbjct: 476 QEQLKLEQIKQEQLKLEQLKQEELKQEQL-KQEQLKQQ 512
Score = 48.5 bits (114), Expect = 5e-05
Identities = 24/101 (23%), Positives = 65/101 (63%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLK 136
++++E ++ +Q++L+Q +++ ++L+QL+ ++ +++ ++ +QL+LKQ +++ LKQ +LK
Sbjct: 383 QLKQEQIKQEQLKLEQIKQEQLKLEQLKQEELKQEQLKQEQLKLKQLKQEE--LKQEQLK 440
Query: 135 HEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNERASI 13
EQ +E+ Q + ++ L +E + ++ E+ +
Sbjct: 441 QEQIKLEQLKQEQLKQEQLKQEQLKKEQLKQEQIKQEQLKL 481
Score = 47.8 bits (112), Expect = 9e-05
Identities = 28/89 (31%), Positives = 51/89 (56%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLK 136
+ ++ +LKQ +LKQ E +LKQ ++K + +LKQ +LKQ + +LKQ ++K
Sbjct: 331 QQEKQQQQLKQEQLKQEELKQEQLKQEQIKLEQLKQEQLKQEQLKQEQLKKEQLKQEQIK 390
Query: 135 HEQSFVEEDSIAQRELHDPEEADLIRESI 49
EQ +E+ Q +L ++ +L +E +
Sbjct: 391 QEQLKLEQIKQEQLKLEQLKQEELKQEQL 419
Score = 45.1 bits (105), Expect = 6e-04
Identities = 28/105 (26%), Positives = 62/105 (58%), Gaps = 8/105 (7%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLR--------LKQREEDSV 160
++++E ++L+Q++ ++ +++ ++ +QL+LKQ +++ ++ +QL+ LKQ +
Sbjct: 398 QIKQEQLKLEQLKQEELKQEQLKQEQLKLKQLKQEELKQEQLKQEQIKLEQLKQEQLKQE 457
Query: 159 RLKQVRLKHEQSFVEEDSIAQRELHDPEEADLIRESIFAQRELNE 25
+LKQ +LK EQ E+ Q +L ++ L E + Q EL +
Sbjct: 458 QLKQEQLKKEQLKQEQIKQEQLKLEQIKQEQLKLEQL-KQEELKQ 501
Score = 43.1 bits (100), Expect = 0.002
Identities = 23/92 (25%), Positives = 58/92 (63%), Gaps = 3/92 (3%)
Frame = -3
Query: 315 RMRRESVRLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLR---LKQREEDSVRLKQV 145
++++E ++ +Q++ +Q +++ ++ +QL+L+Q +++ ++L+QL+ LKQ + +LK
Sbjct: 368 QLKQEQLKQEQLKKEQLKQEQIKQEQLKLEQIKQEQLKLEQLKQEELKQEQLKQEQLKLK 427
Query: 144 RLKHEQSFVEEDSIAQRELHDPEEADLIRESI 49
+LK E+ E+ Q +L ++ L +E +
Sbjct: 428 QLKQEELKQEQLKQEQIKLEQLKQEQLKQEQL 459
Score = 39.7 bits (91), Expect = 0.024
Identities = 22/70 (31%), Positives = 44/70 (62%)
Frame = -3
Query: 294 RLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVE 115
+LKQ LKQ + +LKQ ++KQ++E S++ +QL L+Q+ + + +Q + +H+Q +
Sbjct: 493 QLKQEELKQEQLKQEQLKQQQIKQQQEKSIQQQQL-LEQQLLEQQQHQQQQQQHQQLLEQ 551
Query: 114 EDSIAQRELH 85
+ Q++ H
Sbjct: 552 QQQQHQQQQH 561
Score = 38.9 bits (89), Expect = 0.040
Identities = 24/70 (34%), Positives = 41/70 (58%), Gaps = 3/70 (4%)
Frame = -3
Query: 288 KQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQ---SFV 118
+Q +Q+E+ +LKQ +LKQ E +LKQ ++K + +LKQ +LK EQ +
Sbjct: 325 QQQEKQQQEKQQQQLKQEQLKQEELKQEQLKQEQIKLEQLKQEQLKQEQLKQEQLKKEQL 384
Query: 117 EEDSIAQREL 88
+++ I Q +L
Sbjct: 385 KQEQIKQEQL 394
Score = 32.7 bits (73), Expect = 2.9
Identities = 20/72 (27%), Positives = 42/72 (57%), Gaps = 1/72 (1%)
Frame = -3
Query: 243 KQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVEEDSIAQRELHDPE-EAD 67
+Q +Q+E+ +LKQ +LKQ E +LKQ ++K EQ ++++ + Q +L + + +
Sbjct: 325 QQQEKQQQEKQQQQLKQEQLKQEELKQEQLKQEQIKLEQ--LKQEQLKQEQLKQEQLKKE 382
Query: 66 LIRESIFAQREL 31
+++ Q +L
Sbjct: 383 QLKQEQIKQEQL 394
Score = 31.2 bits (69), Expect = 8.4
Identities = 19/74 (25%), Positives = 42/74 (56%)
Frame = -3
Query: 294 RLKQVRLKQREEDSVRLKQLRLKQREEDSVRLKQLRLKQREEDSVRLKQVRLKHEQSFVE 115
+LKQ ++KQ++E S++ +QL +Q E +Q +Q+++ L+Q + +H+Q +
Sbjct: 508 QLKQQQIKQQQEKSIQQQQLLEQQLLE-----QQQHQQQQQQHQQLLEQQQQQHQQQ--Q 560
Query: 114 EDSIAQRELHDPEE 73
Q++ H ++
Sbjct: 561 HQQYQQQQQHQQQQ 574
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,253,742
Number of Sequences: 1393205
Number of extensions: 8825159
Number of successful extensions: 42483
Number of sequences better than 10.0: 566
Number of HSP's better than 10.0 without gapping: 31735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39093
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)