Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
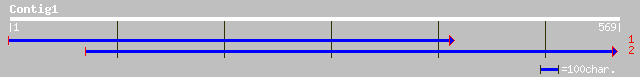
Query= KMC018955A_C01 KMC018955A_c01
(497 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187119.1| unknown protein; protein id: At3g04680.1, suppo... 181 4e-45
ref|NP_198809.1| putative ATP/GTP-binding protein; protein id: A... 114 8e-25
dbj|BAB10217.1| ATP/GTP-binding protein-like [Arabidopsis thaliana] 114 8e-25
gb|EAA04663.1| agCP3430 [Anopheles gambiae str. PEST] 102 3e-21
ref|NP_598601.1| expressed sequence AI462438 [Mus musculus] gi|2... 96 3e-19
>ref|NP_187119.1| unknown protein; protein id: At3g04680.1, supported by cDNA:
gi_17065579 [Arabidopsis thaliana]
gi|6175171|gb|AAF04897.1|AC011437_12 unknown protein
[Arabidopsis thaliana] gi|17065580|gb|AAL32944.1|
Unknown protein [Arabidopsis thaliana]
gi|22136146|gb|AAM91151.1| unknown protein [Arabidopsis
thaliana]
Length = 444
Score = 181 bits (459), Expect = 4e-45
Identities = 83/135 (61%), Positives = 116/135 (85%), Gaps = 1/135 (0%)
Frame = -3
Query: 492 KVRQNARSYKIREYFYGLGNDLSPHSNIANFSDLFVYRIGGGPQAPRSALPIGAEPAADP 313
+VR+ +R++KI+EYFYGL +LSP++N ++FSDL V+RIGGGPQAP+SALP G+ ++P
Sbjct: 309 EVRKRSRNFKIQEYFYGLSKELSPYANTSSFSDLQVFRIGGGPQAPKSALPAGSTSVSNP 368
Query: 312 TRLVPVNIN-HDLLHAVLAVSFAKEPDEIISSNVAGFIYVTDIDIQRKKITYLAPSAGEL 136
R+ PVNI+ DLLH+VLAVS+A+EPD+IISSNV+GF+YVT++++Q+KKITYLAPS G L
Sbjct: 369 LRVTPVNIDDRDLLHSVLAVSYAEEPDQIISSNVSGFVYVTEVNVQKKKITYLAPSPGTL 428
Query: 135 PSKYLIIGNITWRET 91
PSK L+ G++ W E+
Sbjct: 429 PSKLLVAGSLAWLES 443
>ref|NP_198809.1| putative ATP/GTP-binding protein; protein id: At5g39930.1
[Arabidopsis thaliana]
Length = 423
Score = 114 bits (284), Expect = 8e-25
Identities = 58/135 (42%), Positives = 89/135 (64%)
Frame = -3
Query: 495 SKVRQNARSYKIREYFYGLGNDLSPHSNIANFSDLFVYRIGGGPQAPRSALPIGAEPAAD 316
S R+ R+ I+ YFYG+ NDL+ ++ FSD+ VYRIG + ++ + D
Sbjct: 292 SDFRKTLRNSNIQNYFYGVTNDLTVYTKTVKFSDVQVYRIGDFRVSGSTS---AHQRGND 348
Query: 315 PTRLVPVNINHDLLHAVLAVSFAKEPDEIISSNVAGFIYVTDIDIQRKKITYLAPSAGEL 136
P ++ V I+ L++ VLA+S+A +PD+IISS VAGF+ + ++DI ++ITY++PSA EL
Sbjct: 349 PLKITLVTIDEHLVNKVLAISYAIKPDQIISSIVAGFVCIKNVDISEERITYVSPSAAEL 408
Query: 135 PSKYLIIGNITWRET 91
PSK LI+G +TW T
Sbjct: 409 PSKILILGTLTWHVT 423
>dbj|BAB10217.1| ATP/GTP-binding protein-like [Arabidopsis thaliana]
Length = 424
Score = 114 bits (284), Expect = 8e-25
Identities = 58/135 (42%), Positives = 89/135 (64%)
Frame = -3
Query: 495 SKVRQNARSYKIREYFYGLGNDLSPHSNIANFSDLFVYRIGGGPQAPRSALPIGAEPAAD 316
S R+ R+ I+ YFYG+ NDL+ ++ FSD+ VYRIG + ++ + D
Sbjct: 293 SDFRKTLRNSNIQNYFYGVTNDLTVYTKTVKFSDVQVYRIGDFRVSGSTS---AHQRGND 349
Query: 315 PTRLVPVNINHDLLHAVLAVSFAKEPDEIISSNVAGFIYVTDIDIQRKKITYLAPSAGEL 136
P ++ V I+ L++ VLA+S+A +PD+IISS VAGF+ + ++DI ++ITY++PSA EL
Sbjct: 350 PLKITLVTIDEHLVNKVLAISYAIKPDQIISSIVAGFVCIKNVDISEERITYVSPSAAEL 409
Query: 135 PSKYLIIGNITWRET 91
PSK LI+G +TW T
Sbjct: 410 PSKILILGTLTWHVT 424
>gb|EAA04663.1| agCP3430 [Anopheles gambiae str. PEST]
Length = 431
Score = 102 bits (253), Expect = 3e-21
Identities = 54/133 (40%), Positives = 83/133 (61%), Gaps = 1/133 (0%)
Frame = -3
Query: 486 RQNARSYKIREYFYGLGNDLSPHSNIANFSDLFVYRIGGGPQAPRSALPIGAEPAADPTR 307
R AR +IREYFYG L PHS FSD+ ++++G P P S LP+G + + T+
Sbjct: 299 RTEARDQRIREYFYGSKMPLFPHSFDVKFSDIKIFKVGS-PPLPDSCLPLGMKAEDNYTK 357
Query: 306 LVPVNINHDLLHAVLAVSFAKEPDE-IISSNVAGFIYVTDIDIQRKKITYLAPSAGELPS 130
LV V LLH +LAVSFA+ DE +I +NVAGFI VT++++ ++ +T L+P LP
Sbjct: 358 LVAVQPGPQLLHHILAVSFAESTDENVIQTNVAGFICVTNVNMDKQVLTVLSPQPRPLPQ 417
Query: 129 KYLIIGNITWRET 91
L++ ++ + ++
Sbjct: 418 TILLVSDLQFMDS 430
>ref|NP_598601.1| expressed sequence AI462438 [Mus musculus]
gi|27701800|ref|XP_230276.1| similar to expressed
sequence AI462438 [Mus musculus] [Rattus norvegicus]
gi|13096874|gb|AAH03237.1| Similar to ATP/GTP-binding
protein [Mus musculus] gi|26346553|dbj|BAC36924.1|
unnamed protein product [Mus musculus]
Length = 425
Score = 95.5 bits (236), Expect = 3e-19
Identities = 50/132 (37%), Positives = 80/132 (59%), Gaps = 1/132 (0%)
Frame = -3
Query: 486 RQNARSYKIREYFYGLGNDLSPHSNIANFSDLFVYRIGGGPQAPRSALPIGAEPAADPTR 307
R+ R +IREYFYG PH+ FSD+ +Y++G P P S LP+G + +
Sbjct: 293 RRECRDERIREYFYGFRGCFYPHAFNVKFSDVKIYKVGA-PTIPDSCLPLGMSQEDNQLK 351
Query: 306 LVPVNINHDLLHAVLAVSFAKEPDEIIS-SNVAGFIYVTDIDIQRKKITYLAPSAGELPS 130
LVPV D++H +L+VS A+ +E +S ++VAGFI VT +D++ + T L+P+ LP
Sbjct: 352 LVPVTPGRDMVHHLLSVSTAEGTEENLSETSVAGFIVVTSVDVEHQVFTVLSPAPRPLPK 411
Query: 129 KYLIIGNITWRE 94
+L+I +I + +
Sbjct: 412 NFLLIMDIRFMD 423
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 410,499,634
Number of Sequences: 1393205
Number of extensions: 8725600
Number of successful extensions: 26544
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 25735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26511
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 14783057727
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)