Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC018927A_C01 KMC018927A_c01
(620 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
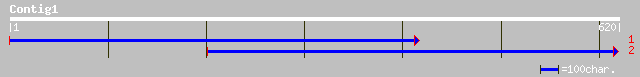
gb|AAM77645.1|AF517846_1 hypothetical protein [Arabidopsis thali... 105 4e-22
ref|NP_172822.1| unknown protein; protein id: At1g13650.1 [Arabi... 64 1e-09
ref|NP_178475.1| hypothetical protein; protein id: At2g03810.1 [... 59 4e-08
gb|AAF14825.1|AC011664_7 hypothetical protein [Arabidopsis thali... 36 0.35
ref|NP_566162.1| hypothetical protein; protein id: At3g02125.1 [... 36 0.35
>gb|AAM77645.1|AF517846_1 hypothetical protein [Arabidopsis thaliana]
Length = 439
Score = 105 bits (263), Expect = 4e-22
Identities = 53/82 (64%), Positives = 64/82 (77%), Gaps = 2/82 (2%)
Frame = -3
Query: 564 GESSFSAVGPVL--SHISYSGSIPYSGSISLRSDSSTTSTRSFAFPVLQNEWNSSPIRMA 391
GE+SFSA V HI+YSG I YSGS+S+RSD+STTS RSFAFP+LQ+EWNSSP+RMA
Sbjct: 359 GETSFSAADSVSISGHITYSGPIAYSGSLSVRSDASTTSGRSFAFPILQSEWNSSPVRMA 418
Query: 390 KADRSRYPKHRSWRERLLCCKF 325
KAD+ R + WR LLCC+F
Sbjct: 419 KADKRR--QKGGWRHTLLCCRF 438
>ref|NP_172822.1| unknown protein; protein id: At1g13650.1 [Arabidopsis thaliana]
gi|25402763|pir||G86269 protein F21F23.9 [imported] -
Arabidopsis thaliana gi|8920570|gb|AAF81292.1|AC027656_9
F21F23.9 [Arabidopsis thaliana]
Length = 281
Score = 64.3 bits (155), Expect = 1e-09
Identities = 32/77 (41%), Positives = 47/77 (60%)
Frame = -3
Query: 603 DDQAFTTEIRHSLGESSFSAVGPVLSHISYSGSIPYSGSISLRSDSSTTSTRSFAFPVLQ 424
D+ + + GE SF L +I+Y G + S ++S+RSD TS SFA P+LQ
Sbjct: 108 DNNLLKRNLPNGFGEISFCEAKSSLDYITYCGPLSGSENLSIRSDG--TSASSFALPILQ 165
Query: 423 NEWNSSPIRMAKADRSR 373
+EWNSSP+RM KA+ ++
Sbjct: 166 SEWNSSPVRMGKAEETQ 182
>ref|NP_178475.1| hypothetical protein; protein id: At2g03810.1 [Arabidopsis
thaliana] gi|25411036|pir||E84452 hypothetical protein
At2g03810 [imported] - Arabidopsis thaliana
gi|4582429|gb|AAD24815.1| hypothetical protein
[Arabidopsis thaliana] gi|20197901|gb|AAM15306.1|
hypothetical protein [Arabidopsis thaliana]
Length = 431
Score = 59.3 bits (142), Expect = 4e-08
Identities = 31/47 (65%), Positives = 37/47 (77%), Gaps = 2/47 (4%)
Frame = -3
Query: 564 GESSFSAVGPVL--SHISYSGSIPYSGSISLRSDSSTTSTRSFAFPV 430
GE+SFSA V HI+YSG I YSGS+S+RSD+STTS RSFAFP+
Sbjct: 369 GETSFSAADSVSISGHITYSGPIAYSGSLSVRSDASTTSGRSFAFPM 415
>gb|AAF14825.1|AC011664_7 hypothetical protein [Arabidopsis thaliana]
Length = 480
Score = 36.2 bits (82), Expect = 0.35
Identities = 24/60 (40%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Frame = -3
Query: 504 IPYSGSISLRSDSSTTSTRSFAFPVLQNE--WNSSPIRMAKADRS--RYPKHRSWRERLL 337
+ +S SIS RSD+S S SFAFP+LQ E +P K + S R P+++ + R+L
Sbjct: 361 VSHSPSISCRSDTSNNSIGSFAFPLLQKEDAVTKTPSMQIKGNVSHKRRPEYQLPQSRML 420
>ref|NP_566162.1| hypothetical protein; protein id: At3g02125.1 [Arabidopsis
thaliana]
Length = 355
Score = 36.2 bits (82), Expect = 0.35
Identities = 24/60 (40%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Frame = -3
Query: 504 IPYSGSISLRSDSSTTSTRSFAFPVLQNE--WNSSPIRMAKADRS--RYPKHRSWRERLL 337
+ +S SIS RSD+S S SFAFP+LQ E +P K + S R P+++ + R+L
Sbjct: 236 VSHSPSISCRSDTSNNSIGSFAFPLLQKEDAVTKTPSMQIKGNVSHKRRPEYQLPQSRML 295
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,194,770
Number of Sequences: 1393205
Number of extensions: 10083186
Number of successful extensions: 26752
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 25960
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26742
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)