Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC018538A_C01 KMC018538A_c01
(948 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
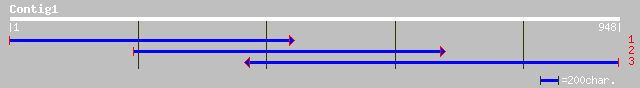
ref|NP_175875.1| hypothetical protein; protein id: At1g54770.1 [... 206 3e-52
pir||G96589 T22H22.18 [imported] - Arabidopsis thaliana gi|37765... 160 3e-38
ref|NP_680266.1| hypothetical protein; protein id: At5g30495.1 [... 146 5e-34
ref|NP_594087.1| hypothetical protein [Schizosaccharomyces pombe... 115 7e-25
emb|CAB92796.1| dJ561L24.2 (acidic 82 kDa protein) [Homo sapiens] 111 1e-23
>ref|NP_175875.1| hypothetical protein; protein id: At1g54770.1 [Arabidopsis
thaliana] gi|27754411|gb|AAO22654.1| unknown protein
[Arabidopsis thaliana] gi|28393935|gb|AAO42375.1|
unknown protein [Arabidopsis thaliana]
Length = 189
Score = 206 bits (525), Expect = 3e-52
Identities = 109/195 (55%), Positives = 140/195 (70%)
Frame = -1
Query: 861 KKPVIGLTWQPQLPISSSLKATDGSQIKTRTEASSSTVWKPNSELVDGLFVPPNDPRKLN 682
K+P+IGL W+P+LP SL GS TR++ + S+ SELVDGL +PPNDPRK+N
Sbjct: 5 KQPLIGLKWEPKLP-GLSLDLKTGS---TRSKTAESS----KSELVDGLCLPPNDPRKIN 56
Query: 681 KLLRKQVKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKKCDSKSKTLPKYF 502
K++RKQ+KDT G NWF+MPA T+TPEL++DL+LLKLR +DP HYKK S+SK KYF
Sbjct: 57 KMIRKQLKDTTGSNWFDMPAPTMTPELKRDLQLLKLRTVMDPALHYKKSVSRSKLAEKYF 116
Query: 501 QMGTVVDSPLDFFSGRLTKAERKASFAEELLSDPNLEAYRKRKVREIEEPNQPAGNENWK 322
Q+GTV++ P + F GRLTK RKA+ A+EL+SDP YRKRKVREIEE ++ N+ W
Sbjct: 117 QIGTVIE-PAEEFYGRLTKKNRKATLADELVSDPKTALYRKRKVREIEEKSRAVTNKKWN 175
Query: 321 IKGGSSRKRAKEKRN 277
KG S K K +RN
Sbjct: 176 KKGNQS-KNTKPRRN 189
>pir||G96589 T22H22.18 [imported] - Arabidopsis thaliana
gi|3776571|gb|AAC64888.1| T22H22.18 [Arabidopsis
thaliana]
Length = 284
Score = 160 bits (404), Expect = 3e-38
Identities = 87/165 (52%), Positives = 112/165 (67%), Gaps = 6/165 (3%)
Frame = -1
Query: 861 KKPVIGLTWQPQLPISSSLKATDGSQIKTRTEASSSTVWKPNSELVDGLFVPPNDPRKLN 682
K+P+IGL W+P+LP SL GS TR++ + S+ SELVDGL +PPNDPRK+N
Sbjct: 5 KQPLIGLKWEPKLP-GLSLDLKTGS---TRSKTAESS----KSELVDGLCLPPNDPRKIN 56
Query: 681 KLLRKQVKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKKCDSKSKTLPKYF 502
K++RKQ+KDT G NWF+MPA T+TPEL++DL+LLKLR +DP HYKK S+SK KYF
Sbjct: 57 KMIRKQLKDTTGSNWFDMPAPTMTPELKRDLQLLKLRTVMDPALHYKKSVSRSKLAEKYF 116
Query: 501 QMG------TVVDSPLDFFSGRLTKAERKASFAEELLSDPNLEAY 385
Q + S + F GRLTK RKA+ A+EL+SDP Y
Sbjct: 117 QASFRTRSKRITVSFAEEFYGRLTKKNRKATLADELVSDPKTALY 161
>ref|NP_680266.1| hypothetical protein; protein id: At5g30495.1 [Arabidopsis
thaliana]
Length = 166
Score = 146 bits (368), Expect = 5e-34
Identities = 77/163 (47%), Positives = 102/163 (62%)
Frame = -1
Query: 870 MPEKKPVIGLTWQPQLPISSSLKATDGSQIKTRTEASSSTVWKPNSELVDGLFVPPNDPR 691
M E KP+IGLTW+P+LP S T + K SS++W SELVDGL +PPNDP+
Sbjct: 1 MAETKPLIGLTWEPKLPGLSLDTKTCSTSSKRVESHESSSLWMSKSELVDGLCLPPNDPK 60
Query: 690 KLNKLLRKQVKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKKCDSKSKTLP 511
K+NK++RKQ+KDT G NWF+MPA T+TPEL++DL+LLK+
Sbjct: 61 KINKMIRKQIKDTTGSNWFDMPAPTMTPELKRDLQLLKI--------------------- 99
Query: 510 KYFQMGTVVDSPLDFFSGRLTKAERKASFAEELLSDPNLEAYR 382
GTV++ P + F GRLTK RKA+ A+EL+SDP + YR
Sbjct: 100 -----GTVIE-PAEEFYGRLTKKNRKATLADELVSDPKVSQYR 136
>ref|NP_594087.1| hypothetical protein [Schizosaccharomyces pombe]
gi|7491110|pir||T11651 hypothetical protein SPAC3G9.15c
- fission yeast (Schizosaccharomyces pombe)
gi|2706466|emb|CAA15924.1| eukarotic conserved protein;
similar to S. cerevisiae YLR051C [Schizosaccharomyces
pombe]
Length = 230
Score = 115 bits (289), Expect = 7e-25
Identities = 50/117 (42%), Positives = 80/117 (67%)
Frame = -1
Query: 672 RKQVKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKKCDSKSKTLPKYFQMG 493
+K +KDTAG NWF+MPA +T +++D++LLK+R A+DPKRHY++ +K++PKYFQ+G
Sbjct: 104 KKNIKDTAGSNWFDMPATELTESVKRDIQLLKMRNALDPKRHYRR--ENTKSMPKYFQVG 161
Query: 492 TVVDSPLDFFSGRLTKAERKASFAEELLSDPNLEAYRKRKVREIEEPNQPAGNENWK 322
++V+ P DF+S R+ ERK + +ELL D +Y K+K E+++ +K
Sbjct: 162 SIVEGPQDFYSSRIPTRERKETIVDELLHDSERRSYFKKKYLELQKSKMSGRKGQYK 218
>emb|CAB92796.1| dJ561L24.2 (acidic 82 kDa protein) [Homo sapiens]
Length = 756
Score = 111 bits (278), Expect = 1e-23
Identities = 63/136 (46%), Positives = 81/136 (59%), Gaps = 4/136 (2%)
Frame = -1
Query: 711 VPPNDPRK--LNKLLRKQVKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKK 538
VPP K L K RK+ + TAG WF M A +T EL+ DLK LK+R ++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 537 CDSKSKTLPKYFQMGTVVDSPLDFFSGRLTKAERKASFAEELLSDPNLEAYRKRKVREI- 361
D PKYFQ+GT+VD+P DF+ R+ K +RK + EELL+D Y +RK EI
Sbjct: 679 NDRDG--FPKYFQIGTIVDNPADFYHSRIPKKQRKRTIVEELLADSEFRRYNRRKYSEIM 736
Query: 360 -EEPNQPAGNENWKIK 316
E+ AG + K K
Sbjct: 737 AEKAANAAGKKFRKKK 752
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 846,760,547
Number of Sequences: 1393205
Number of extensions: 19321348
Number of successful extensions: 56839
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 53468
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56785
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53246406144
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)